
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,749,416 – 13,749,691 |
| Length | 275 |
| Max. P | 0.998574 |

| Location | 13,749,416 – 13,749,532 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -27.92 |
| Energy contribution | -28.28 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13749416 116 + 22407834 CA----ACCGGAUUCUGACUGAAAUCCAUUUUACGGGUGCCAAGUGCCAGACCAAACCGAAUGUCUGUAAACUUGCACGUGACUUUGGCGCAACUUGCAAUAUUUGCAGCUACGGCCAAA ..----...((((((.....).)))))......((.((((.((((..(((((..........)))))...)))))))).))...(((((.....(((((.....))))).....))))). ( -31.00) >DroSec_CAF1 13053 116 + 1 CA----ACCGUAUUCUGACUGAAAUCCAUUUUACGGGUGCCAAGUGCCAGACCAAACCGAAUGUCUGUAAACUUGCACGUGAGUUUGGCGCAACUUGCAAUAUUUGCAGCUACGGCCAAA ..----.(((((...((.........))...)))))((((.((((..(((((..........)))))...)))))))).....((((((.....(((((.....))))).....)))))) ( -29.60) >DroAna_CAF1 24871 116 + 1 CAGGAUGCUGGAUG----CUGAAAUCCAUUUUACGGGUGCCAAGUGCCAGACCAAACCAAAUGUCUGUAAACUUGCACGUGACUUUGGCCCAACUUGCAAUAUUUGCAGCUAAGGCUAAA ........(((((.----.....))))).....((.((((.((((..(((((..........)))))...)))))))).))...((((((....(((((.....)))))....)))))). ( -32.60) >DroSim_CAF1 15082 116 + 1 CA----ACCGUAUUCUGACUGAAAUCCAUUUUACGGGUGCCAAGUGCCAGACCAAACCGAAUGUCUGUAAACUUGCACGUGAGUUUGGCGCAACUUGCAAUAUUUGCAGCUACGGCCAAA ..----.(((((...((.........))...)))))((((.((((..(((((..........)))))...)))))))).....((((((.....(((((.....))))).....)))))) ( -29.60) >DroEre_CAF1 15109 112 + 1 CA----ACCGGAUUCU----GAAAUCCAUUUUACGGGUGCCAAGUGCCAGACAAAACCGAAUGUCUGUAAACUUGCACGUGACUUUGGCGCAACUUGCAAUAUUUGCAGCUAUGGCCAAA ..----...(((((..----..)))))......((.((((.((((..((((((........))))))...)))))))).))...(((((.((..(((((.....)))))...))))))). ( -34.20) >DroYak_CAF1 15890 112 + 1 CG----ACCGGAUUCU----GAAAUCCAUUUUACGGGUGCCAAGUGCCAGACCAAACCGAAUGUCUGUAAACUUGCACGUGACUUUGGCGCAACUUGCAAUAUUUGCAGCUAAGGCCAAA ..----...(((((..----..)))))......((.((((.((((..(((((..........)))))...)))))))).))...(((((.....(((((.....))))).....))))). ( -30.60) >consensus CA____ACCGGAUUCU__CUGAAAUCCAUUUUACGGGUGCCAAGUGCCAGACCAAACCGAAUGUCUGUAAACUUGCACGUGACUUUGGCGCAACUUGCAAUAUUUGCAGCUACGGCCAAA .........(((((........)))))......((.((((.((((..(((((..........)))))...)))))))).))..((((((.....(((((.....))))).....)))))) (-27.92 = -28.28 + 0.36)


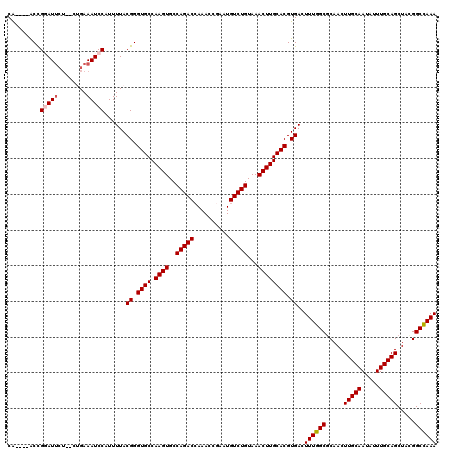
| Location | 13,749,416 – 13,749,532 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -30.60 |
| Energy contribution | -31.47 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13749416 116 - 22407834 UUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUCACGUGCAAGUUUACAGACAUUCGGUUUGGUCUGGCACUUGGCACCCGUAAAAUGGAUUUCAGUCAGAAUCCGGU----UG ((((((.(((((((((.....))).))))))))))))((((.((((.((....(((((.....)))))..)).)))).))))..........(((((((......)))))))..----.. ( -38.90) >DroSec_CAF1 13053 116 - 1 UUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAACUCACGUGCAAGUUUACAGACAUUCGGUUUGGUCUGGCACUUGGCACCCGUAAAAUGGAUUUCAGUCAGAAUACGGU----UG ((((((.(((((((((.....))).)))))))))))).....((((((((...(((((..........)))))..)))).))))(((((.....(((....)))....))))).----.. ( -36.90) >DroAna_CAF1 24871 116 - 1 UUUAGCCUUAGCUGCAAAUAUUGCAAGUUGGGCCAAAGUCACGUGCAAGUUUACAGACAUUUGGUUUGGUCUGGCACUUGGCACCCGUAAAAUGGAUUUCAG----CAUCCAGCAUCCUG ....((((.(((((((.....))).))))))))....((((.((((.((....(((((.....)))))..)).)))).))))..........(((((.....----.)))))........ ( -31.30) >DroSim_CAF1 15082 116 - 1 UUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAACUCACGUGCAAGUUUACAGACAUUCGGUUUGGUCUGGCACUUGGCACCCGUAAAAUGGAUUUCAGUCAGAAUACGGU----UG ((((((.(((((((((.....))).)))))))))))).....((((((((...(((((..........)))))..)))).))))(((((.....(((....)))....))))).----.. ( -36.90) >DroEre_CAF1 15109 112 - 1 UUUGGCCAUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUCACGUGCAAGUUUACAGACAUUCGGUUUUGUCUGGCACUUGGCACCCGUAAAAUGGAUUUC----AGAAUCCGGU----UG ((((((..((((((((.....))).))))).)))))).....((((((((...((((((........))))))..)))).))))........((((((..----..))))))..----.. ( -34.00) >DroYak_CAF1 15890 112 - 1 UUUGGCCUUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUCACGUGCAAGUUUACAGACAUUCGGUUUGGUCUGGCACUUGGCACCCGUAAAAUGGAUUUC----AGAAUCCGGU----CG ((((((..((((((((.....))).))))).))))))((((.((((.((....(((((.....)))))..)).)))).))))..........((((((..----..))))))..----.. ( -33.60) >consensus UUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUCACGUGCAAGUUUACAGACAUUCGGUUUGGUCUGGCACUUGGCACCCGUAAAAUGGAUUUCAG__AGAAUCCGGU____UG ((((((.(((((((((.....))).)))))))))))).....((((((((...(((((..........)))))..)))).))))........(((((((......)))))))........ (-30.60 = -31.47 + 0.86)



| Location | 13,749,452 – 13,749,571 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -25.03 |
| Energy contribution | -24.92 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13749452 119 + 22407834 CAAGUGCCAGACCAAACCGAAUGUCUGUAAACUUGCACGUGACUUUGGCGCAACUUGCAAUAUUUGCAGCUACGGCCAAAACCAAAACUAAGCUGCCAACAACAAUGGCAACUC-CGGCC (((((..(((((..........)))))...)))))..((.((.((((((.....(((((.....))))).....)))))).............(((((.......)))))..))-))... ( -26.90) >DroSec_CAF1 13089 120 + 1 CAAGUGCCAGACCAAACCGAAUGUCUGUAAACUUGCACGUGAGUUUGGCGCAACUUGCAAUAUUUGCAGCUACGGCCAAAACCAAAACUAAGCUGCCAACAACAAUGGCAACUCCCGGCC (((((..(((((..........)))))...)))))..((.(((((((((.....(((((.....))))).....)))))..............(((((.......))))))))).))... ( -30.20) >DroAna_CAF1 24907 117 + 1 CAAGUGCCAGACCAAACCAAAUGUCUGUAAACUUGCACGUGACUUUGGCCCAACUUGCAAUAUUUGCAGCUAAGGCUAAAACCAAAACUAAGCUGCCAACAACAAUGGGAGC---GGAGC (((((..(((((..........)))))...)))))....((..(((((((....(((((.....)))))....)))))))..))...((..(((.(((.......))).)))---..)). ( -30.60) >DroSim_CAF1 15118 120 + 1 CAAGUGCCAGACCAAACCGAAUGUCUGUAAACUUGCACGUGAGUUUGGCGCAACUUGCAAUAUUUGCAGCUACGGCCAAAACCAAAACUAAGCUGCCAACAACAAUGGCAACUCCCGGCC (((((..(((((..........)))))...)))))..((.(((((((((.....(((((.....))))).....)))))..............(((((.......))))))))).))... ( -30.20) >DroEre_CAF1 15141 119 + 1 CAAGUGCCAGACAAAACCGAAUGUCUGUAAACUUGCACGUGACUUUGGCGCAACUUGCAAUAUUUGCAGCUAUGGCCAAAACCAAAACUAAGCUGCCAACAACAAUGGCGACUCCUGG-A (((((..((((((........))))))...)))))....((..((((((.((..(((((.....)))))...))))))))..))...(((((.(((((.......))))).))..)))-. ( -30.20) >DroYak_CAF1 15922 119 + 1 CAAGUGCCAGACCAAACCGAAUGUCUGUAAACUUGCACGUGACUUUGGCGCAACUUGCAAUAUUUGCAGCUAAGGCCAAAACCAAAACUAAGCUGCCAACAACAAUGGCAACUCCUGG-A (((((..(((((..........)))))...)))))....((.(((((((((((..........)))).))))))).))...(((......((.(((((.......))))).))..)))-. ( -27.50) >consensus CAAGUGCCAGACCAAACCGAAUGUCUGUAAACUUGCACGUGACUUUGGCGCAACUUGCAAUAUUUGCAGCUACGGCCAAAACCAAAACUAAGCUGCCAACAACAAUGGCAACUCCCGGCC (((((..(((((..........)))))...)))))....((..((((((.....(((((.....))))).....))))))..)).........(((((.......))))).......... (-25.03 = -24.92 + -0.11)


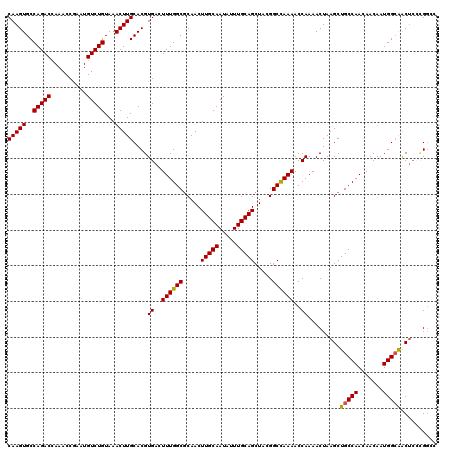
| Location | 13,749,452 – 13,749,571 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -41.43 |
| Consensus MFE | -38.04 |
| Energy contribution | -39.07 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13749452 119 - 22407834 GGCCG-GAGUUGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUCACGUGCAAGUUUACAGACAUUCGGUUUGGUCUGGCACUUG .((((-(((((((((.......)))))))...((..((..((((((.(((((((((.....))).))))))))))))..))...)).......(((((.....))))).))))))..... ( -46.20) >DroSec_CAF1 13089 120 - 1 GGCCGGGAGUUGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAACUCACGUGCAAGUUUACAGACAUUCGGUUUGGUCUGGCACUUG .((((((((((((((.......))))))))).((..((..((((((.(((((((((.....))).))))))))))))..))...)).......(((((.....)))))..)))))..... ( -45.10) >DroAna_CAF1 24907 117 - 1 GCUCC---GCUCCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUAGCCUUAGCUGCAAAUAUUGCAAGUUGGGCCAAAGUCACGUGCAAGUUUACAGACAUUUGGUUUGGUCUGGCACUUG (((..---(((.(((.......))).)))..)))..((..(((.((((.(((((((.....))).)))))))).)))..)).((((.((....(((((.....)))))..)).))))... ( -33.20) >DroSim_CAF1 15118 120 - 1 GGCCGGGAGUUGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAACUCACGUGCAAGUUUACAGACAUUCGGUUUGGUCUGGCACUUG .((((((((((((((.......))))))))).((..((..((((((.(((((((((.....))).))))))))))))..))...)).......(((((.....)))))..)))))..... ( -45.10) >DroEre_CAF1 15141 119 - 1 U-CCAGGAGUCGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCAUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUCACGUGCAAGUUUACAGACAUUCGGUUUUGUCUGGCACUUG .-....((((.((((.......)))).)))).....((..((((((..((((((((.....))).))))).))))))..))....(((((...((((((........))))))..))))) ( -38.10) >DroYak_CAF1 15922 119 - 1 U-CCAGGAGUUGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCUUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUCACGUGCAAGUUUACAGACAUUCGGUUUGGUCUGGCACUUG .-....(((((((((.......))))))))).....((..((((((..((((((((.....))).))))).))))))..)).((((.((....(((((.....)))))..)).))))... ( -40.90) >consensus GGCCGGGAGUUGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUCACGUGCAAGUUUACAGACAUUCGGUUUGGUCUGGCACUUG ......(((((((((.......))))))))).....((..((((((.(((((((((.....))).))))))))))))..)).((((.((....(((((.....)))))..)).))))... (-38.04 = -39.07 + 1.03)


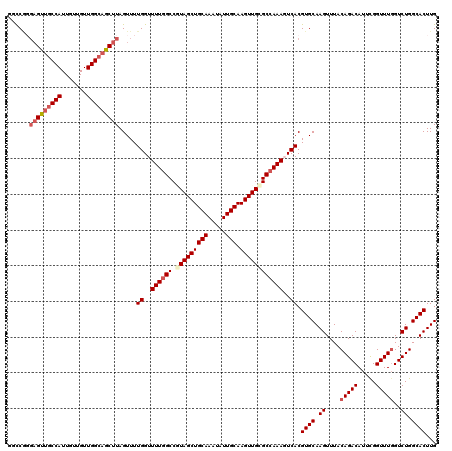
| Location | 13,749,492 – 13,749,611 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -31.60 |
| Energy contribution | -32.47 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13749492 119 - 22407834 AUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUUGGCCG-GAGUUGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUC ...(((...(((((((.....)))))))..))).((.((((((..-(((((((((.......)))))))))......(((....)))(((((((((.....))).)))))))))))).)) ( -40.10) >DroSec_CAF1 13129 120 - 1 AUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUUGGCCGGGAGUUGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAACUC ...(((...(((((((.....)))))))..))).(((((((((...(((((((((.......)))))))))......(((....)))(((((((((.....))).))))))))))))))) ( -44.00) >DroAna_CAF1 24947 117 - 1 AUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUUGCUCC---GCUCCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUAGCCUUAGCUGCAAAUAUUGCAAGUUGGGCCAAAGUC ....((((((((((((.....))))))).....((((....))))---((((...((((((((.((((((.......(((....)))..)))))).))))..))))...))))..))))) ( -32.90) >DroSim_CAF1 15158 120 - 1 AUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUUGGCCGGGAGUUGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAACUC ...(((...(((((((.....)))))))..))).(((((((((...(((((((((.......)))))))))......(((....)))(((((((((.....))).))))))))))))))) ( -44.00) >DroEre_CAF1 15181 119 - 1 AUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUUU-CCAGGAGUCGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCAUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUC ....(((..((((((((...((((..((((((.((......-)).))))))((((.......)))))))).)))))))).((((((..((((((((.....))).))))).))))))))) ( -34.50) >DroYak_CAF1 15962 119 - 1 AUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUUU-CCAGGAGUUGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCUUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUC ...(((...(((((((.....)))))))..)))((......-))..(((((((((.......)))))))))......(..((((((..((((((((.....))).))))).))))))..) ( -35.20) >consensus AUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUUGGCCGGGAGUUGCCAUUGUUGUUGGCAGCUUAGUUUUGGUUUUGGCCGUAGCUGCAAAUAUUGCAAGUUGCGCCAAAGUC .(.(((...(((((((.....)))))))..))).)...........(((((((((.......)))))))))......(..((((((.(((((((((.....))).))))))))))))..) (-31.60 = -32.47 + 0.86)

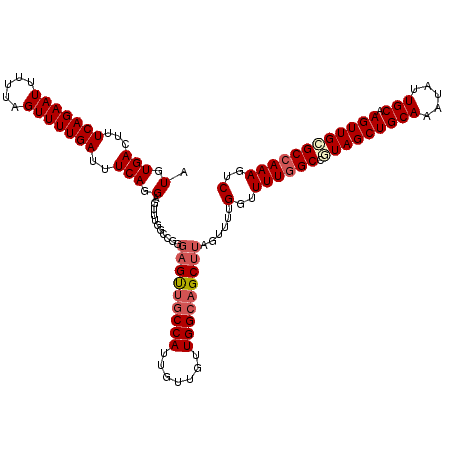

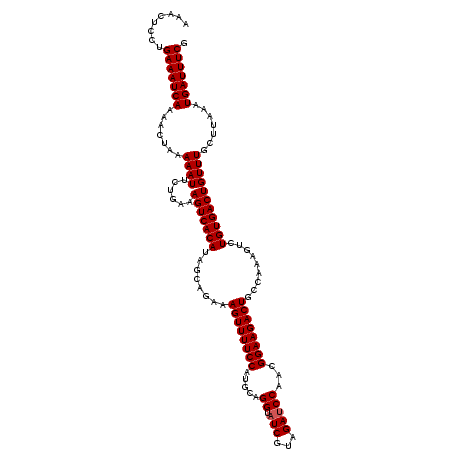
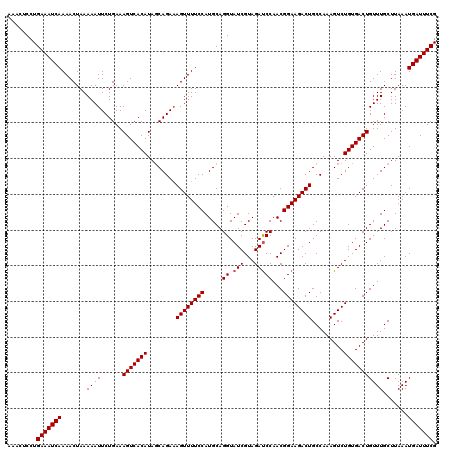
| Location | 13,749,571 – 13,749,691 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -23.75 |
| Energy contribution | -23.92 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13749571 120 + 22407834 AAACUCCUGAAAUCAAAACUAAAAAUUCUGAAAGUCACAUAGCAGAAAGUUUUCCAUGCAGGUAUCGUAGAUCCAACGGAAGACUGCCAAAGUCUGUGACUGUUUGCUUAAAUGAUUUCG ........(((((((.......((((......(((((((........((((((((.....((.(((...)))))...)))))))).........))))))))))).......))))))). ( -24.57) >DroSec_CAF1 13209 120 + 1 AAACUCCUGAAAUCAAAACUAAAAAUUCUGAAAGUCACAUAGCAGAAAGUUUUCCAUGCAGGUAUCGUAGAUCCAACGGAAGACUGCCAAAGUCUGUGACUGUUUGCUUAAAUGAUUUCG ........(((((((.......((((......(((((((........((((((((.....((.(((...)))))...)))))))).........))))))))))).......))))))). ( -24.57) >DroAna_CAF1 25024 120 + 1 AAACUCCUGAAAUCAAAACUAAAAAUUCUGAAAGUCACAUACCAGAAAGUUUUCCAUGCAGGUAUCGUAGACCCAACGGAAGACUGCCAAAGUCUGUGACUGUUUGCUUAAAUGAUUUCG ........(((((((.......((((......(((((((.((.....((((((((.....(((.......)))....))))))))......)).))))))))))).......))))))). ( -27.14) >DroSim_CAF1 15238 120 + 1 AAACUCCUGAAAUCAAAACUAAAAAUUCUGAAAGUCACAUAGCAGAAAGUUUUCCAUGCAGGUAUCGUAGAUCCAACGGAAGACUGCCAAAGUCUGUGACUGUUUGCUUAAAUGAUUUCG ........(((((((.......((((......(((((((........((((((((.....((.(((...)))))...)))))))).........))))))))))).......))))))). ( -24.57) >DroEre_CAF1 15260 120 + 1 AAACUCCUGAAAUCAAAACUAAAAAUUCUGAAAGUCACAUAGCAGAAAGUUUUCCAUGCAGGUAUCGUAGAUCCAACGGAAGACUGCCAAAGUCUGUGACUGUUUGCUUAAAUGAUUUCG ........(((((((.......((((......(((((((........((((((((.....((.(((...)))))...)))))))).........))))))))))).......))))))). ( -24.57) >DroYak_CAF1 16041 119 + 1 AAACUCCUGAAAUCAAAACUAAAAAUUCUGAAAGUCACAUAGCAGA-AGUUUUCCAUGCAGGUAUCGUAGAUCCAACGGAAGACUGCCAAAGUCUGUGACUGUUUGCUUAAAUGAUUUCG ........(((((((.......((((......(((((((.......-((((((((.....((.(((...)))))...)))))))).........))))))))))).......))))))). ( -24.13) >consensus AAACUCCUGAAAUCAAAACUAAAAAUUCUGAAAGUCACAUAGCAGAAAGUUUUCCAUGCAGGUAUCGUAGAUCCAACGGAAGACUGCCAAAGUCUGUGACUGUUUGCUUAAAUGAUUUCG ........(((((((.......((((......(((((((........((((((((.....((.(((...)))))...)))))))).........))))))))))).......))))))). (-23.75 = -23.92 + 0.17)

| Location | 13,749,571 – 13,749,691 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -26.34 |
| Energy contribution | -26.20 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
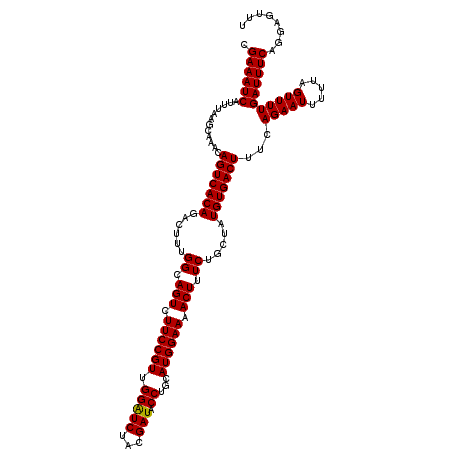
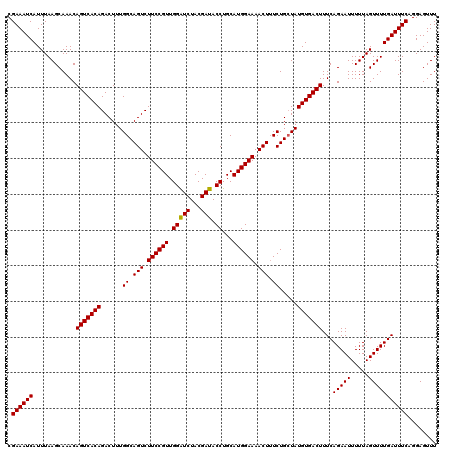
>2L_DroMel_CAF1 13749571 120 - 22407834 CGAAAUCAUUUAAGCAAACAGUCACAGACUUUGGCAGUCUUCCGUUGGAUCUACGAUACCUGCAUGGAAAACUUUCUGCUAUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUU .((((((............(((((((.....((((((..((((((.(((((...))).))...))))))......)))))))))))))...(((((.....)))))))))))........ ( -27.30) >DroSec_CAF1 13209 120 - 1 CGAAAUCAUUUAAGCAAACAGUCACAGACUUUGGCAGUCUUCCGUUGGAUCUACGAUACCUGCAUGGAAAACUUUCUGCUAUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUU .((((((............(((((((.....((((((..((((((.(((((...))).))...))))))......)))))))))))))...(((((.....)))))))))))........ ( -27.30) >DroAna_CAF1 25024 120 - 1 CGAAAUCAUUUAAGCAAACAGUCACAGACUUUGGCAGUCUUCCGUUGGGUCUACGAUACCUGCAUGGAAAACUUUCUGGUAUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUU .((((((............(((((((.(((..((.(((.(((((((((((.......))))).)))))).))).)).))).)))))))...(((((.....)))))))))))........ ( -30.80) >DroSim_CAF1 15238 120 - 1 CGAAAUCAUUUAAGCAAACAGUCACAGACUUUGGCAGUCUUCCGUUGGAUCUACGAUACCUGCAUGGAAAACUUUCUGCUAUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUU .((((((............(((((((.....((((((..((((((.(((((...))).))...))))))......)))))))))))))...(((((.....)))))))))))........ ( -27.30) >DroEre_CAF1 15260 120 - 1 CGAAAUCAUUUAAGCAAACAGUCACAGACUUUGGCAGUCUUCCGUUGGAUCUACGAUACCUGCAUGGAAAACUUUCUGCUAUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUU .((((((............(((((((.....((((((..((((((.(((((...))).))...))))))......)))))))))))))...(((((.....)))))))))))........ ( -27.30) >DroYak_CAF1 16041 119 - 1 CGAAAUCAUUUAAGCAAACAGUCACAGACUUUGGCAGUCUUCCGUUGGAUCUACGAUACCUGCAUGGAAAACU-UCUGCUAUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUU .((((((............(((((((.....((((((..((((((.(((((...))).))...))))))....-.)))))))))))))...(((((.....)))))))))))........ ( -27.90) >consensus CGAAAUCAUUUAAGCAAACAGUCACAGACUUUGGCAGUCUUCCGUUGGAUCUACGAUACCUGCAUGGAAAACUUUCUGCUAUGUGACUUUCAGAAUUUUUAGUUUUGAUUUCAGGAGUUU .((((((............(((((((......((.(((.((((((.(((((...))).))...)))))).))).)).....)))))))...(((((.....)))))))))))........ (-26.34 = -26.20 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:34 2006