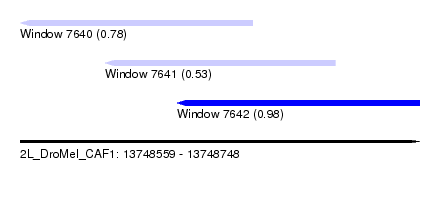
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,748,559 – 13,748,748 |
| Length | 189 |
| Max. P | 0.979467 |

| Location | 13,748,559 – 13,748,669 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 91.13 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -22.90 |
| Energy contribution | -24.71 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13748559 110 - 22407834 ACACGUGCACAUAUACAGAACAUUCAAUGUGUGUGUGUGUGUGUGGAGGAGCAUCUUGUGGCUGGUU--UCCUGGUCCAAUGUUGACUGCUUGUUUUUAUUCCAUUUUUUUU (((((..((((((((((..........))))))))))..)))))(((((((((((...((((..(..--..)..).))).....)).))))).......))))......... ( -35.11) >DroSec_CAF1 12243 107 - 1 ACACGUGCACAUAUACAGAGCAUUCAAUGUCUGU----GUGUGUGGAGGAGCAUCUUGUGGCUGGUU-UUCCUUGUCCAAUGUUGACUGCUUGUUUUUAUUCCAUUUUUUUU .......((((((((((((.(((...))))))))----)))))))((.(((((((...((((.((..-..))..).))).....)).))))).))................. ( -28.00) >DroSim_CAF1 14272 107 - 1 ACACGUGCACAUAUACAGAGCAUUCAAUGUCUGU----GUGUGUGGAGGAGCAUCUUGUGGCUGGUU-UUCCUUGUCCAAUGUUGACUGCUUGUUUUUAUUCCAUUUUUUUU .......((((((((((((.(((...))))))))----)))))))((.(((((((...((((.((..-..))..).))).....)).))))).))................. ( -28.00) >DroEre_CAF1 14135 109 - 1 ACACGUCCACACAUACAGGCCAUUGCAUGUGUGUGUGAGUGUGUGGAGGAGCAUCUUGUGGCUGGUUUUUCCUUGUCCAAUGUUGACUGCUUGUUUUUAUUCCAUUUUU--- (((((..((((((((((.((....)).))))))))))..)))))(((((((((((...((((.((.....))..).))).....)).))))).......))))......--- ( -35.41) >consensus ACACGUGCACAUAUACAGAGCAUUCAAUGUCUGU____GUGUGUGGAGGAGCAUCUUGUGGCUGGUU_UUCCUUGUCCAAUGUUGACUGCUUGUUUUUAUUCCAUUUUUUUU (((((..((((((((((..........))))))))))..)))))(((((((((((...((((.((.....))..).))).....)).))))).......))))......... (-22.90 = -24.71 + 1.81)

| Location | 13,748,599 – 13,748,708 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.49 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -23.76 |
| Energy contribution | -25.82 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13748599 109 - 22407834 UCAAGUGUGAAGGUGUGCGUGUGUGUGUGUGUGGUCGUUACACGUGCACAUAUACAGAACAUUCAAUGUGUGUGUGUGUGUGUGGAGGAGCAUCUUGUGGCUGGUU--UCC ...(((...((((((((((..(....)..)))..((.(((((((..((((((((((..........))))))))))..)))))))..)))))))))...)))....--... ( -33.90) >DroSec_CAF1 12283 102 - 1 UCAAGUGUGAAGGUGUGCGUGUGU----GUGUGCUCGUUACACGUGCACAUAUACAGAGCAUUCAAUGUCUGU----GUGUGUGGAGGAGCAUCUUGUGGCUGGUU-UUCC ....((((((.((((..((....)----)..))))..))))))...((((((((((((.(((...))))))))----)))))))(((((.((((....)).)).))-))). ( -32.70) >DroSim_CAF1 14312 102 - 1 UCAAGUGUGAAGGUGUGCGUGUGU----GUGUGCUCGUUACACGUGCACAUAUACAGAGCAUUCAAUGUCUGU----GUGUGUGGAGGAGCAUCUUGUGGCUGGUU-UUCC ....((((((.((((..((....)----)..))))..))))))...((((((((((((.(((...))))))))----)))))))(((((.((((....)).)).))-))). ( -32.70) >DroEre_CAF1 14172 105 - 1 UCAAGUGUGAAGGUGUGCGUGU------GUGUGCUCGUUACACGUCCACACAUACAGGCCAUUGCAUGUGUGUGUGAGUGUGUGGAGGAGCAUCUUGUGGCUGGUUUUUCC ........(((((...((....------..((((((.(((((((..((((((((((.((....)).))))))))))..)))))))..))))))......))....))))). ( -37.90) >consensus UCAAGUGUGAAGGUGUGCGUGUGU____GUGUGCUCGUUACACGUGCACAUAUACAGAGCAUUCAAUGUCUGU____GUGUGUGGAGGAGCAUCUUGUGGCUGGUU_UUCC ...(((...((((((((((..(......)..)))((.(((((((..((((((((((..........))))))))))..)))))))..)))))))))...)))......... (-23.76 = -25.82 + 2.06)


| Location | 13,748,633 – 13,748,748 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -36.42 |
| Energy contribution | -37.48 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13748633 115 - 22407834 ACGGAUGUUGAAUUGGUUUGCCUUCGUCUCGGCUUGCCACUCAAGUGUGAAGGUGUGCGUGUGUGUGUGUGUGGUCGUUACACGUGCACAUAUACAGAACAUUCAAUGUGUGUGU (((.(((((((((.(((.((((((((.(...(((((.....)))))))))))))).))(((((((((..((((.......))))..)))))))))....)))))))))).))).. ( -43.00) >DroSec_CAF1 12316 109 - 1 ACGGAUGUUGAAUUGGUUUGCCUUCGUCUCGGCUUGCCACUCAAGUGUGAAGGUGUGCGUGUGU----GUGUGCUCGUUACACGUGCACAUAUACAGAGCAUUCAAUGUCUGU-- (((((((((((((((((..(((........)))..))))(((..(..(......)..).(((((----((((((.((.....)).)))))))))))))).)))))))))))))-- ( -44.90) >DroSim_CAF1 14345 109 - 1 ACGGAUGUUGAAUUGGUUUGCCUUCGUCUCGGCUUGCCACUCAAGUGUGAAGGUGUGCGUGUGU----GUGUGCUCGUUACACGUGCACAUAUACAGAGCAUUCAAUGUCUGU-- (((((((((((((((((..(((........)))..))))(((..(..(......)..).(((((----((((((.((.....)).)))))))))))))).)))))))))))))-- ( -44.90) >DroEre_CAF1 14208 109 - 1 ACGGAUGUUGAAUUGGUUUGCCUUCGUCUCGGCUUGCCACUCAAGUGUGAAGGUGUGCGUGU------GUGUGCUCGUUACACGUCCACACAUACAGGCCAUUGCAUGUGUGUGU ..((((.((((..((((..(((........)))..)))).))))((((((.((((..(....------)..))))..))))))))))(((((((((.((....)).))))))))) ( -40.80) >consensus ACGGAUGUUGAAUUGGUUUGCCUUCGUCUCGGCUUGCCACUCAAGUGUGAAGGUGUGCGUGUGU____GUGUGCUCGUUACACGUGCACAUAUACAGAGCAUUCAAUGUCUGU__ (((((((((((((((((..(((........)))..)))).....(((((...(((..((((((...............))))))..))).))))).....))))))))))))).. (-36.42 = -37.48 + 1.06)

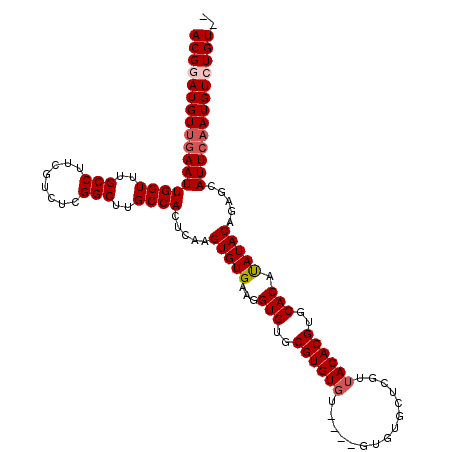
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:27 2006