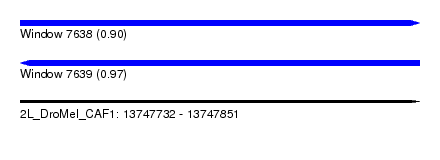
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,747,732 – 13,747,851 |
| Length | 119 |
| Max. P | 0.968935 |

| Location | 13,747,732 – 13,747,851 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.79 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13747732 119 + 22407834 CUUAUGUGUUGACUUUAACUGGAGAUUCUUAAAGUAGUACAUCAAAAGGUUGAUUUGCAUGUUGAGAACAACCCUUUAAUGGAUACUACUUAAGGAAGCUUCCAUCAACAAUUU-UUCUU ......((((((.......(((((.(((((.(((((((((((.(((.(((((.(((........))).))))).))).)))..)))))))).)))))..)))))))))))....-..... ( -30.61) >DroSec_CAF1 11409 119 + 1 UUUA-AUGUUGACUUUGACUGUAGAUUCCUAAAGUAGUACAUCAAACGGUUGAUUUGCAUGUUGAGAACAACCCUUUAAUGGAUACUACUUAAGGAAGCUUCCAUGAACAAUUUUUUCUU ....-.((((.((.......))((.(((((.(((((((((((.(((.(((((.(((........))).))))).))).)))..)))))))).))))).))......)))).......... ( -23.90) >DroAna_CAF1 23266 104 + 1 AUUU-UAGCCGACUUUAAAUGGAGCUACCUAAAGUAGUACAUCAAACGGUCACUUUGAAU-AUAUUAACGACCCUUUAAUGGAUACUACUUAAGGAAACUGCCAUA-------------- ....-.............((((((...(((.(((((((((((.(((.((((...((((..-...)))).)))).))).)))..)))))))).)))...)).)))).-------------- ( -22.80) >DroSim_CAF1 13126 119 + 1 UUUA-AUGUUGACUUUGACUGUAGAUUCCUAAAGUAGUACAUCAAACGGUUGAUUUGCAUGUUGAGAACAACCCUUUAAUGGAUACUACUUAAGGAAGCUUCCAUGAACAAUUUUUUCUU ....-.((((.((.......))((.(((((.(((((((((((.(((.(((((.(((........))).))))).))).)))..)))))))).))))).))......)))).......... ( -23.90) >DroEre_CAF1 13197 117 + 1 AUUA-UUGUUGACUUUGACUGGAGAUUCUUAAAGUAGUACAUUAAACGGUCGAUUCA-AUUGUGACCACAACCCUUUAAUGGAUACUACUUAAGGAAGCUUCCAUCAACAACUU-UUCUU ....-..((((...((((.(((((.(((((.(((((((((((((((.(((.(..(((-....)))...).))).)))))))..)))))))).)))))..)))))))))))))..-..... ( -31.50) >DroYak_CAF1 14003 117 + 1 AUUA-AUGUUGACUUUGACUGGAGAUUCCUAAAGUAGUAAAUCAAAAGGUUGAUCUU-AUUGUGACCACAACCCUUUAAUGGAUACUACUUAAGGAAGCUUCCAUCAAACACUU-UUCCU ....-........(((((.(((((.(((((.((((((((.((.(((.(((((.((..-.....))...))))).))).))...)))))))).)))))..)))))))))).....-..... ( -29.80) >consensus AUUA_AUGUUGACUUUGACUGGAGAUUCCUAAAGUAGUACAUCAAACGGUUGAUUUGCAUGUUGAGAACAACCCUUUAAUGGAUACUACUUAAGGAAGCUUCCAUCAACAAUUU_UUCUU ...................(((((.(((((.(((((((((((.(((.(((((................))))).))).)))..)))))))).)))))..)))))................ (-21.07 = -21.79 + 0.72)



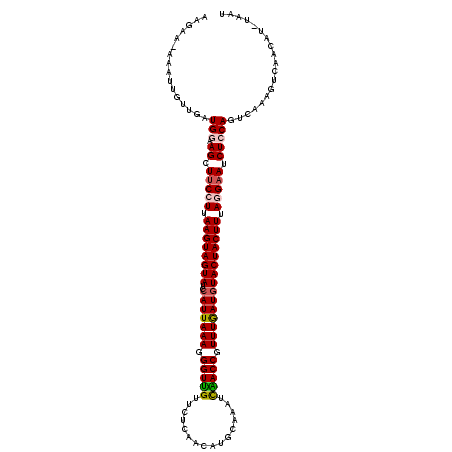
| Location | 13,747,732 – 13,747,851 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -26.69 |
| Energy contribution | -27.06 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.22 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13747732 119 - 22407834 AAGAA-AAAUUGUUGAUGGAAGCUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUUCUCAACAUGCAAAUCAACCUUUUGAUGUACUACUUUAAGAAUCUCCAGUUAAAGUCAACACAUAAG .....-....((((((((((.(.(((...((((((((..(((((((((((((................)))))))))))))))))))))...))).))))).......))))))...... ( -32.40) >DroSec_CAF1 11409 119 - 1 AAGAAAAAAUUGUUCAUGGAAGCUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUUCUCAACAUGCAAAUCAACCGUUUGAUGUACUACUUUAGGAAUCUACAGUCAAAGUCAACAU-UAAA ..((....(((((....(((...(((((.((((((((..(((((((.(((((................))))).))))))))))))))).))))))))))))).....)).....-.... ( -30.09) >DroAna_CAF1 23266 104 - 1 --------------UAUGGCAGUUUCCUUAAGUAGUAUCCAUUAAAGGGUCGUUAAUAU-AUUCAAAGUGACCGUUUGAUGUACUACUUUAGGUAGCUCCAUUUAAAGUCGGCUA-AAAU --------------.((((.((((.(((.((((((((..(((((((.(((((.......-........))))).))))))))))))))).))).)))))))).............-.... ( -32.46) >DroSim_CAF1 13126 119 - 1 AAGAAAAAAUUGUUCAUGGAAGCUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUUCUCAACAUGCAAAUCAACCGUUUGAUGUACUACUUUAGGAAUCUACAGUCAAAGUCAACAU-UAAA ..((....(((((....(((...(((((.((((((((..(((((((.(((((................))))).))))))))))))))).))))))))))))).....)).....-.... ( -30.09) >DroEre_CAF1 13197 117 - 1 AAGAA-AAGUUGUUGAUGGAAGCUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUGGUCACAAU-UGAAUCGACCGUUUAAUGUACUACUUUAAGAAUCUCCAGUCAAAGUCAACAA-UAAU .....-..((((((((((((.(.(((...((((((((..(((((((.(((((...(((....-)))..))))).)))))))))))))))...))).))))).......)))))))-)... ( -36.31) >DroYak_CAF1 14003 117 - 1 AGGAA-AAGUGUUUGAUGGAAGCUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUGGUCACAAU-AAGAUCAACCUUUUGAUUUACUACUUUAGGAAUCUCCAGUCAAAGUCAACAU-UAAU .....-.(((((((((((((.(.(((((.((((((((...((((((((((..(((((.....-..))))))))))))))).)))))))).))))).))))).))).....)))))-)... ( -37.70) >consensus AAGAA_AAAUUGUUGAUGGAAGCUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUUCUCAACAUGCAAAUCAACCGUUUGAUGUACUACUUUAGGAAUCUCCAGUCAAAGUCAACAU_UAAU ................(((.((.(((((.((((((((..(((((((.(((((................))))).))))))))))))))).))))).)))))................... (-26.69 = -27.06 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:24 2006