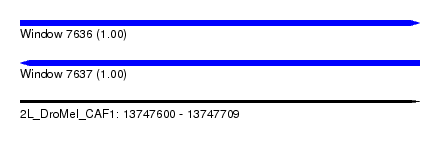
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,747,600 – 13,747,709 |
| Length | 109 |
| Max. P | 0.997807 |

| Location | 13,747,600 – 13,747,709 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.30 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -14.80 |
| Energy contribution | -15.12 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13747600 109 + 22407834 GUUGGGAUACAAAAUCAGCUAGUUAAGUAGGUCAUACAAUGCGACACUUUUAUAUCAAAUCGCCCUUUGUAUCCACUACUUAAAUAUAUAAUUUCUAAUUAAUUCCGAU---------- ((((((((..............((((((((...((((((.((((...............))))...))))))...)))))))).....((((.....))))))))))))---------- ( -21.26) >DroSec_CAF1 11257 119 + 1 GUUGGAAUACAAAAUCAGCUAGUUAAGUAGGCCAUACAAUGCGAUACUUUUAUAUCAAAUCGCCUUUUGUAUCCACUACUUAAAUAUAUAAUUUCUAAAUAUUUCCGAUUAUUAUUAUA (((((((....((((.......((((((((...((((((.(((((.............)))))...))))))...)))))))).......))))........))))))).......... ( -21.76) >DroAna_CAF1 23127 104 + 1 GCUUGCA---AAAAUCAACUAGUUAUGUAGGCUAUACAUUGCGAGUAUAUUUUAUUUAGUCGCCC-AUGUAUUGCCUACUUAAACACAUGAUUUAG--UUUGCACAGAU---------A .((((((---(((((((.....(((.((((((.((((((.((((.((.........)).))))..-)))))).)))))).))).....)))))...--)))))).))..---------. ( -31.10) >DroSim_CAF1 12974 119 + 1 GUUGGAAUACAAAAUCAGCUAGUUAAGUAGGCCAUACAAUGCGAUACUUUUAUAUCAAAUCGCCUUUUGUAUCCACUACUUAAAUAUAUAAUUUCUAAAUAUUUCCGAUUAUUAUUAUA (((((((....((((.......((((((((...((((((.(((((.............)))))...))))))...)))))))).......))))........))))))).......... ( -21.76) >DroEre_CAF1 13082 101 + 1 GUUGGGAUACAAAAUCAGCUAGUUAAGCAGGUCAUACAAUGCGACAUUUUUAUACCAAAUCGCCGUUUGUAUCCACUACUUAGAUAUAUAAUUUUGAUCAA------------------ .....(((.((((((.......(((((.((...((((((.((((...............))))...))))))...)).))))).......)))))))))..------------------ ( -17.10) >consensus GUUGGAAUACAAAAUCAGCUAGUUAAGUAGGCCAUACAAUGCGACACUUUUAUAUCAAAUCGCCCUUUGUAUCCACUACUUAAAUAUAUAAUUUCUAAAUAUUUCCGAU_________A (((((.........)))))...((((((((...((((((.((((...............))))...))))))...)))))))).................................... (-14.80 = -15.12 + 0.32)

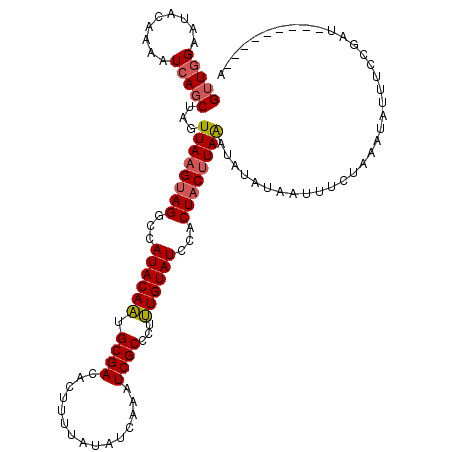
| Location | 13,747,600 – 13,747,709 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.30 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.14 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13747600 109 - 22407834 ----------AUCGGAAUUAAUUAGAAAUUAUAUAUUUAAGUAGUGGAUACAAAGGGCGAUUUGAUAUAAAAGUGUCGCAUUGUAUGACCUACUUAACUAGCUGAUUUUGUAUCCCAAC ----------...(((..((....(((((((..((.((((((((.(.((((((...(((((.............))))).))))))..))))))))).))..))))))).))))).... ( -23.32) >DroSec_CAF1 11257 119 - 1 UAUAAUAAUAAUCGGAAAUAUUUAGAAAUUAUAUAUUUAAGUAGUGGAUACAAAAGGCGAUUUGAUAUAAAAGUAUCGCAUUGUAUGGCCUACUUAACUAGCUGAUUUUGUAUUCCAAC .............((((.(((...(((((((..((.((((((((...((((((...(((((.............))))).))))))...)))))))).))..))))))))))))))... ( -25.02) >DroAna_CAF1 23127 104 - 1 U---------AUCUGUGCAAA--CUAAAUCAUGUGUUUAAGUAGGCAAUACAU-GGGCGACUAAAUAAAAUAUACUCGCAAUGUAUAGCCUACAUAACUAGUUGAUUUU---UGCAAGC .---------.....((((((--...(((((...(((...((((((.((((((-..((((.((.........)).)))).)))))).))))))..)))....)))))))---))))... ( -30.50) >DroSim_CAF1 12974 119 - 1 UAUAAUAAUAAUCGGAAAUAUUUAGAAAUUAUAUAUUUAAGUAGUGGAUACAAAAGGCGAUUUGAUAUAAAAGUAUCGCAUUGUAUGGCCUACUUAACUAGCUGAUUUUGUAUUCCAAC .............((((.(((...(((((((..((.((((((((...((((((...(((((.............))))).))))))...)))))))).))..))))))))))))))... ( -25.02) >DroEre_CAF1 13082 101 - 1 ------------------UUGAUCAAAAUUAUAUAUCUAAGUAGUGGAUACAAACGGCGAUUUGGUAUAAAAAUGUCGCAUUGUAUGACCUGCUUAACUAGCUGAUUUUGUAUCCCAAC ------------------..(((((((((((..((..(((((((.(.((((((...(((((.............))))).))))))..))))))))..))..)))))))).)))..... ( -24.62) >consensus U_________AUCGGAAAUAAUUAGAAAUUAUAUAUUUAAGUAGUGGAUACAAAAGGCGAUUUGAUAUAAAAGUAUCGCAUUGUAUGGCCUACUUAACUAGCUGAUUUUGUAUCCCAAC ........................(((((((..((.((((((((...((((((...(((((.............))))).))))))...)))))))).))..))))))).......... (-20.42 = -20.14 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:22 2006