| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,747,007 – 13,747,131 |
| Length | 124 |
| Max. P | 0.770314 |

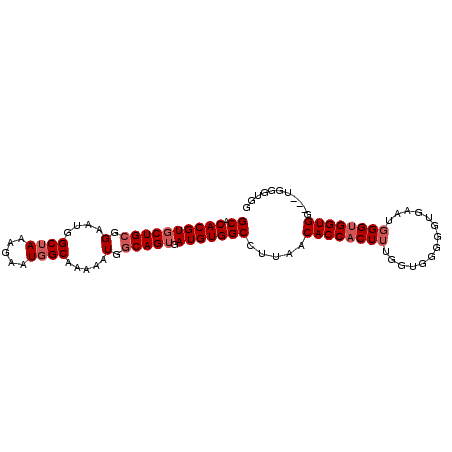
| Location | 13,747,007 – 13,747,102 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.59 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -21.86 |
| Energy contribution | -23.02 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13747007 95 + 22407834 ----CACACACACUUCUCAUUGUCGCAGCUUCGGCACACGUGCUGCGGAAUGGCUAAAGAAUGGCAAAAAUGGCAGUGAUGUGGCCUUAACACCACUUU ----............(((((((((((((..((.....)).)))))......((((.....))))......)))))))).((((........))))... ( -28.10) >DroSec_CAF1 10674 99 + 1 CACACACACACACUUCUCAUUGUCGCAGCUUCGGCACACGUGCUGCGGAAUGGCUAAAGAAUGGCAAAAAUGGCAGUGAUGUGGCCUUAACACCACUUU ................(((((((((((((..((.....)).)))))......((((.....))))......)))))))).((((........))))... ( -28.10) >DroSim_CAF1 12391 99 + 1 CACACACACACACUUCUCAUUGUCGCAGCUUCGGCACACGUGCUGCGGAAUGGCUAAAGAAUGGCAAAAAUGGCAGUGAUGUGGCCUUAACACCACUUU ................(((((((((((((..((.....)).)))))......((((.....))))......)))))))).((((........))))... ( -28.10) >DroEre_CAF1 12534 90 + 1 ---------ACACUUCCCAUUGUCGCAGCUCCGGCACACGUGCUGAAGAAUGGCGAAAGAAUGGCGAAAAUGGCAGGGAUGUGGCCUUAACACCACUUU ---------...(((.(((((.((((..((.(((((....))))).)).....(....)....)))).))))).)))...((((........))))... ( -26.80) >DroYak_CAF1 13366 95 + 1 ----CACACACACUUCCCAUUGUCGCAGCUUCGGCACACGUGCUGAAGAAUGGCGAAAGUAUGGCAGAAAUGGCAGUGAUGUGGCCUUAACUCCACUUU ----..((((((((..((((..((((..((((((((....))))))))....))))....))))((....))..)))).))))................ ( -31.70) >consensus ____CACACACACUUCUCAUUGUCGCAGCUUCGGCACACGUGCUGCGGAAUGGCUAAAGAAUGGCAAAAAUGGCAGUGAUGUGGCCUUAACACCACUUU ..........((((.((((((.(((((((..((.....)).)))))))))))((((.....))))......)).))))..((((........))))... (-21.86 = -23.02 + 1.16)

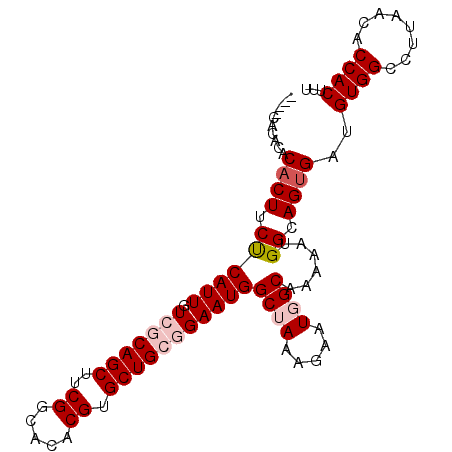
| Location | 13,747,036 – 13,747,131 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -21.72 |
| Energy contribution | -22.97 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13747036 95 + 22407834 GCACACGUGCUGCGGAAUGGCUAAAGAAUGGCAAAAAUGGCAGUGAUGUGGCCUUAACACCACUUUGGUGGGGGUAAGUUGGUGGUGG---UGGGUAG ...((((((((((.(....((((.....)))).....).))))).)))))((((...(((((((....((....))....))))))).---.)))).. ( -31.00) >DroSec_CAF1 10707 95 + 1 GCACACGUGCUGCGGAAUGGCUAAAGAAUGGCAAAAAUGGCAGUGAUGUGGCCUUAACACCACUUUGGUGGGGGUGAAUGGGUGGUGG---UGGGUGG ...((((((((((.(....((((.....)))).....).))))).)))))((((...((((((((..............)))))))).---.)))).. ( -31.64) >DroSim_CAF1 12424 95 + 1 GCACACGUGCUGCGGAAUGGCUAAAGAAUGGCAAAAAUGGCAGUGAUGUGGCCUUAACACCACUUUGGUGGGGGUGAAUGGGUGGUGG---UGGGUGG ...((((((((((.(....((((.....)))).....).))))).)))))((((...((((((((..............)))))))).---.)))).. ( -31.64) >DroEre_CAF1 12558 91 + 1 GCACACGUGCUGAAGAAUGGCGAAAGAAUGGCGAAAAUGGCAGGGAUGUGGCCUUAACACCACUUUGGUGGGGGUGA-UGGGGGGUGCGUG------G ...((((..((...(.....(....).....).....(.(((....))).)(((((.((((.(((....))))))).-)))))))..))))------. ( -25.30) >consensus GCACACGUGCUGCGGAAUGGCUAAAGAAUGGCAAAAAUGGCAGUGAUGUGGCCUUAACACCACUUUGGUGGGGGUGAAUGGGUGGUGG___UGGGUGG ((.((((((((((.(....((((.....)))).....).))))).))))))).....((((((((..............))))))))........... (-21.72 = -22.97 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:20 2006