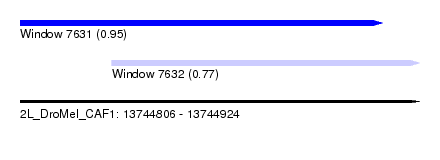
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,744,806 – 13,744,924 |
| Length | 118 |
| Max. P | 0.953134 |

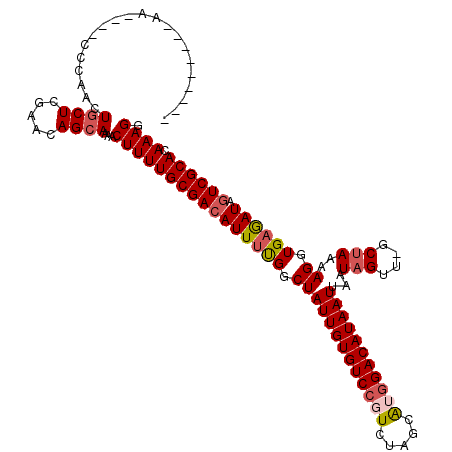
| Location | 13,744,806 – 13,744,913 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.66 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -26.50 |
| Energy contribution | -27.25 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13744806 107 + 22407834 ---------AA----CACAACUGCUCGAACAGCAAAACUUUUGCGACAUUUUGGCUAUUGUGUCCGUCUAGCAAGGACAUAAUUAAUAGUUGGCUAAAAGGUGAGAUAGUCGCACAAAGG ---------..----......((((.....))))...((((((((((((((((.(((((((((((.........)))))))))...(((....)))..)).)))))).)))))).)))). ( -29.70) >DroSec_CAF1 8387 105 + 1 ---------AA------CAACUGCUCGAAUAGCAAAACUUUUGCGACAUUUUGGCUAUUGUGUCCGUCUAGCAUGGACAUAAUUAAUAGUUGGCUAAAAGGUGAGAUAGUCGCACAAAGG ---------..------....((((.....))))...((((((((((((((((.(((((((((((((.....)))))))))))...(((....)))..)).)))))).)))))).)))). ( -31.10) >DroAna_CAF1 20322 119 + 1 AACCGGCCAAAAUGCCCAAAAUGCUCGAACAGAAAAACUUUUGCGACAUUCCGACUAUUGUGUCCGUCUAAUGGAGACAUAAUUAAUAGUU-GCCAAAAGGUGAAAUACUCGCACAAAGG ....(((......))).....................(((((((((......(((((((((.(((((...))))).)))......))))))-(((....))).......))))).)))). ( -24.00) >DroSim_CAF1 10104 105 + 1 ---------AA------CAACUGCUCGAAUAGCAAAACUUUUGCGACAUUUUGGCUAUUGUGUCCGUCUAGCAUGGACAUAAUUAAUAGUUGGCUAAAAGGUGAGAUAGUCGCACAAAGG ---------..------....((((.....))))...((((((((((((((((.(((((((((((((.....)))))))))))...(((....)))..)).)))))).)))))).)))). ( -31.10) >DroEre_CAF1 10068 110 + 1 ---------AAAGGGCCGAAGUGCUCGAACAGCAAAACUUUUGCGACAUUUUGGCUAUUGUGUCCGUCUAGGAUGGACAUAAUUAAUAGUU-GCUAAAAGGUGAGAUAGUCGCACAAAGG ---------...((((......))))...........(((((((((((((((((((((((((((((((...))))))))))....))))))-(((....)))))))).)))))).)))). ( -33.10) >DroYak_CAF1 10460 110 + 1 ---------AAAGGGCCGAACUGCUCGAACAGCAAAACUUUUGCGACAUUUUGGCUAUUGUGUCCGUCUAGCAUGGACAUAAUUAAUAGUU-GCUAAAAGGUGAGAUAGUCGCACAAAGG ---------...((((......))))...........((((((((((((((((((((((((((((((.....)))))))))....))))))-(((....)))))))).)))))).)))). ( -30.90) >consensus _________AA____CCCAACUGCUCGAACAGCAAAACUUUUGCGACAUUUUGGCUAUUGUGUCCGUCUAGCAUGGACAUAAUUAAUAGUU_GCUAAAAGGUGAGAUAGUCGCACAAAGG .....................((((.....))))...((((((((((((((((.(((((((((((((.....)))))))))))...(((....)))..)).)))))).)))))).)))). (-26.50 = -27.25 + 0.75)

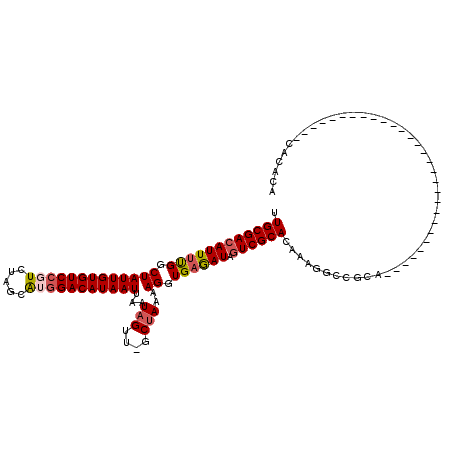
| Location | 13,744,833 – 13,744,924 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.34 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -22.47 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13744833 91 + 22407834 UUGCGACAUUUUGGCUAUUGUGUCCGUCUAGCAAGGACAUAAUUAAUAGUUGGCUAAAAGGUGAGAUAGUCGCACAAAGGCCGCA-----------------------------CACAUA .((((((((((((.(((((((((((.........)))))))))...(((....)))..)).)))))).))))))...........-----------------------------...... ( -24.60) >DroSec_CAF1 8412 91 + 1 UUGCGACAUUUUGGCUAUUGUGUCCGUCUAGCAUGGACAUAAUUAAUAGUUGGCUAAAAGGUGAGAUAGUCGCACAAAGGCCGCA-----------------------------CACACA .((((((((((((.(((((((((((((.....)))))))))))...(((....)))..)).)))))).))))))...........-----------------------------...... ( -26.10) >DroAna_CAF1 20362 119 + 1 UUGCGACAUUCCGACUAUUGUGUCCGUCUAAUGGAGACAUAAUUAAUAGUU-GCCAAAAGGUGAAAUACUCGCACAAAGGUCGCCAUGUGUGUGCCCCACACCUGGCAUUGUUCCAUGGA .((.((((....((((.((((((..((((.....))))...........(.-.((....))..).......)))))).))))((((.(((((.....))))).))))..)))).)).... ( -35.70) >DroSim_CAF1 10129 91 + 1 UUGCGACAUUUUGGCUAUUGUGUCCGUCUAGCAUGGACAUAAUUAAUAGUUGGCUAAAAGGUGAGAUAGUCGCACAAAGGCCGCA-----------------------------CACACA .((((((((((((.(((((((((((((.....)))))))))))...(((....)))..)).)))))).))))))...........-----------------------------...... ( -26.10) >DroEre_CAF1 10099 90 + 1 UUGCGACAUUUUGGCUAUUGUGUCCGUCUAGGAUGGACAUAAUUAAUAGUU-GCUAAAAGGUGAGAUAGUCGCACAAAGGCCGCA-----------------------------CACACA .((((...(((((((.((((((((((((...))))))))))))........-))))))).((((.....))))........))))-----------------------------...... ( -26.50) >DroYak_CAF1 10491 90 + 1 UUGCGACAUUUUGGCUAUUGUGUCCGUCUAGCAUGGACAUAAUUAAUAGUU-GCUAAAAGGUGAGAUAGUCGCACAAAGGCCGCA-----------------------------UACACA .((((...(((((((.(((((((((((.....)))))))))))........-))))))).((((.....))))........))))-----------------------------...... ( -24.40) >consensus UUGCGACAUUUUGGCUAUUGUGUCCGUCUAGCAUGGACAUAAUUAAUAGUU_GCUAAAAGGUGAGAUAGUCGCACAAAGGCCGCA_____________________________CACACA .((((((((((((.(((((((((((((.....)))))))))))...(((....)))..)).)))))).)))))).............................................. (-22.47 = -23.05 + 0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:18 2006