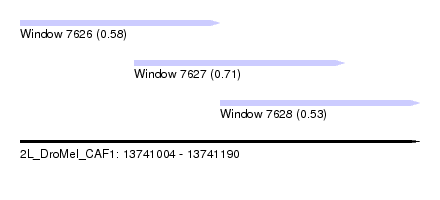
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,741,004 – 13,741,190 |
| Length | 186 |
| Max. P | 0.713854 |

| Location | 13,741,004 – 13,741,097 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -21.15 |
| Energy contribution | -21.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13741004 93 + 22407834 UCAAAGCGGCC---------------------------AACUCCACCCACUCCACCCACACAUUGGCGUGUGGUGAGGCGUGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUG .......(((.---------------------------.....(((((((.(((.........))).))).))))((((((((.(((....)))..))))))))......)))....... ( -28.00) >DroSec_CAF1 4785 84 + 1 UCAAAGCCACC---------------------------C---------ACUCCGCCCACACAUUGGCGUGUGGUGAGGCGUGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUG .....((((((---------------------------.---------((..((((........)))).))))))((((((((.(((....)))..)))))))))).............. ( -24.50) >DroAna_CAF1 17000 82 + 1 UCAAGGAC---------------------------------CUCG-----CCAGCUCACACUUUGGCGUGUGGUGAGGCGUGUUAUCGUCGGAUUUGCAUGCUUGCACAAGUCAUAAAUG .....(((---------------------------------(.((-----((((........)))))).)..(((((((((((.(((....)))..)))))))).)))..)))....... ( -25.20) >DroSim_CAF1 6443 93 + 1 UCAAAGCCACC---------------------------CACUCUACCCACUCCGCCCACACAUUGGCGUGUGGUGAGGCGUGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUG .....((....---------------------------..(((...((((..((((........)))).)))).)))((((((.(((....)))..))))))..)).............. ( -24.80) >DroEre_CAF1 6307 93 + 1 UCAAAACCGUC---------------------------CACUCCGCCCACUACGCCCACACAUUGGCGUGUGGUGAGGCGUGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUG .((((.(((.(---------------------------....((((((((((((((((.....))).)))))))).)))).).....).))).))))...((((....))))........ ( -31.40) >DroYak_CAF1 6533 120 + 1 UCAAAACCACCCACUCCGCCCACUCCGCCCACUCCGCCCACUCCGCCCACUUCGCCCACACAUUGGCGUGUGGUGAGGCGUGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUG ..........................((....(((((.....(((((((((.((((((.....))).))).)))).)))).).....).))))...))..((((....))))........ ( -27.50) >consensus UCAAAGCCACC___________________________CACUCCGCCCACUCCGCCCACACAUUGGCGUGUGGUGAGGCGUGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUG ........................................................(((((......)))))(((((((((((.(((....)))..)))))))).)))............ (-21.15 = -21.15 + 0.00)

| Location | 13,741,057 – 13,741,155 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.29 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -23.09 |
| Energy contribution | -22.52 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13741057 98 + 22407834 UGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUGUCAAUUGGCAUGGCA---------GGCGGAAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGU------------- .....((.(((((......(((((((.((.((((...........)))).)))))---------))))....))))).))(((....)))(((....))).......------------- ( -34.80) >DroSec_CAF1 4829 98 + 1 UGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUGUCAAUUGGCAUGGCA---------GGCGGGAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGC------------- .....((.(((((.((...(((((((.((.((((...........)))).)))))---------)))).)).))))).))(((....)))(((....))).......------------- ( -35.00) >DroAna_CAF1 17042 115 + 1 UGUUAUCGUCGGAUUUGCAUGCUUGCACAAGUCAUAAAUGUCAAUUGGUGGGGCAACUCCAGCAGCCGG-----CGAAGGGGCAAGUGGUCGGCCAUCUACGUAAGUGGCAAGUAUGUGG ..............(..(((((((((....(((.....((((..(((.((((....).))).)))..))-----))....)))..((((....)))).(((....))))))))))))..) ( -35.60) >DroSim_CAF1 6496 98 + 1 UGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUGUCAAUUGGCAUGGCA---------GGCGGGAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGU------------- .....((.(((((.((...(((((((.((.((((...........)))).)))))---------)))).)).))))).))(((....)))(((....))).......------------- ( -35.00) >DroEre_CAF1 6360 111 + 1 UGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUGUCAAUUGGCAUGGCA---------AGCGGGAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGUGCCGAGGAAAUGG .....((.(((((((((...((((((.((.((((...........)))).)))))---------)))(...(((((...((((....)))).)))))...).))))).)))).))..... ( -38.50) >DroYak_CAF1 6613 111 + 1 UGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUGUCAAUUGGCAUGGCA---------GGCGGGAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGUGCCGAGGAAAUGG .....((.(((((((((...((((((.((.((((...........)))).)))))---------)))(...(((((...((((....)))).)))))...).))))).)))).))..... ( -38.20) >consensus UGUUAUCGUCGGAUUUGCAUGCUUGCACAAGCCAUAAAUGUCAAUUGGCAUGGCA_________GGCGGGAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGU_____________ ......(.(((((..((((....))))...((((....((((....))))))))..................))))).)((((....))))((....))..................... (-23.09 = -22.52 + -0.58)
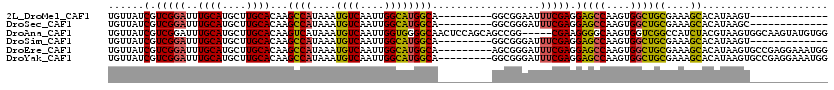
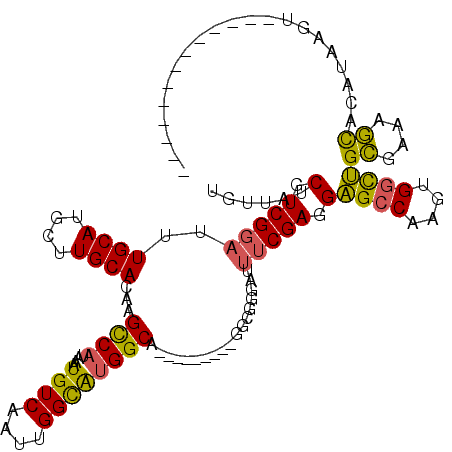
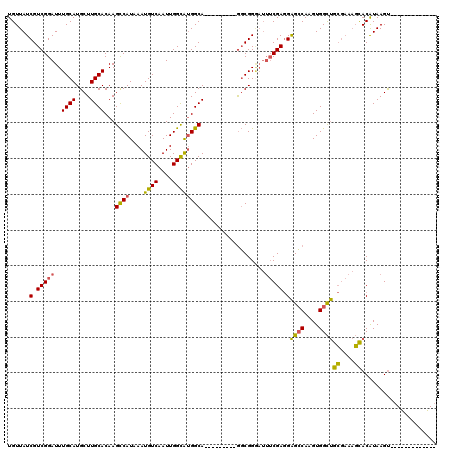
| Location | 13,741,097 – 13,741,190 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.16 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -26.34 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13741097 93 + 22407834 UCAAUUGGCAUGGCAGGCGGAAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGU------------------GCCCCAGCUGGUGUCAUAAUUACAGAUGACUAUCA ...((..((.(((..((((...........((((....))))((....)).......)------------------))))))))..))(((((........)))))..... ( -28.00) >DroSec_CAF1 4869 93 + 1 UCAAUUGGCAUGGCAGGCGGGAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGC------------------GCCCCAGCUGGUGUCAUAAUUACAGAUGACUAUCA ...((..((.(((..((((...........((((....))))((....)).......)------------------))))))))..))(((((........)))))..... ( -30.00) >DroSim_CAF1 6536 93 + 1 UCAAUUGGCAUGGCAGGCGGGAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGU------------------GCCCCAGCUGGUGUCAUAAUUACAGAUGACUAUCA (((.(((..((((((((((((.(((((...((((....)))).)))))((((....))------------------))))).)))..)))))).....))).)))...... ( -29.70) >DroEre_CAF1 6400 111 + 1 UCAAUUGGCAUGGCAAGCGGGAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGUGCCGAGGAAAUGGAAGGCGCCCCAGCUGGCGUCAUAAUUACAGAUGACUAUCA (((.(((.(((..(...(((.((((.....((((....))))((....))....)))).)))..)..)))...((((((......)))))).......))).)))...... ( -33.10) >DroYak_CAF1 6653 111 + 1 UCAAUUGGCAUGGCAGGCGGGAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGUGCCGAGGAAAUGGAAAGCGCCCCAGCUGGUGUCAUAAUUACAGAUGACUAUCA ...((..((.(((..((((...((((..(.((((....)))).)....((((....))))..........)))).)))))))))..))(((((........)))))..... ( -34.30) >consensus UCAAUUGGCAUGGCAGGCGGGAUUUCGAGGAGCCAAGUGGCUGCGAAAGCACAUAAGU__________________GCCCCAGCUGGUGUCAUAAUUACAGAUGACUAUCA ...((..((.(((..(((............((((....))))((....))..........................))))))))..))(((((........)))))..... (-26.34 = -26.74 + 0.40)

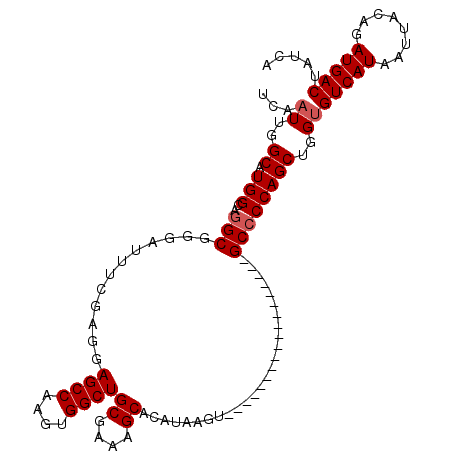
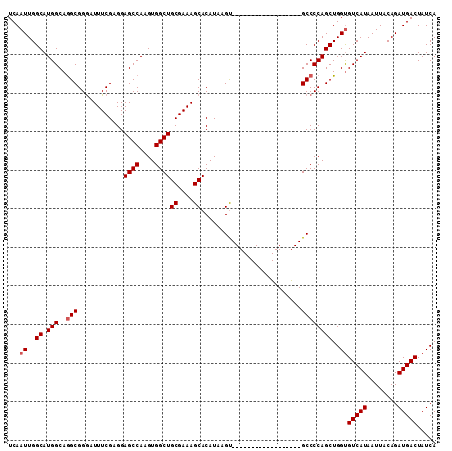
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:14 2006