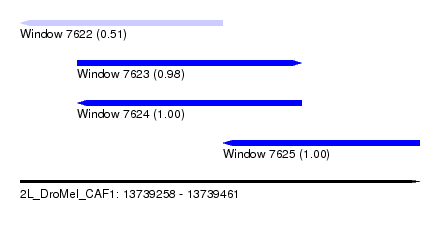
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,739,258 – 13,739,461 |
| Length | 203 |
| Max. P | 0.998053 |

| Location | 13,739,258 – 13,739,361 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 85.26 |
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -14.82 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
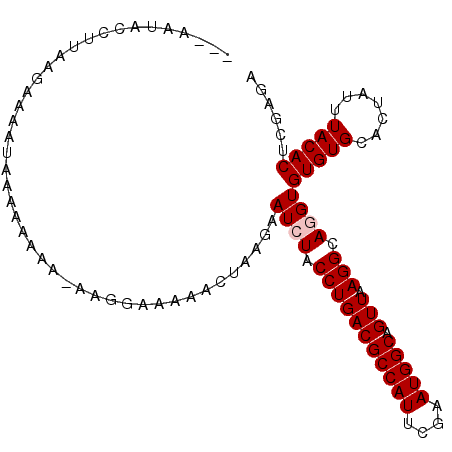
>2L_DroMel_CAF1 13739258 103 - 22407834 AAAAAUACGUUAAGAAAAUAAAAAAAA-AAGGAAAAACUAAGGAUCUACCUGACGCCAUUCGAAUGGCAGUUAAGGCAGGUGUGUGCACUAUUUACACUCGAGA .............((............-...............((((.(((((((((((....))))).))).))).))))(((((.......))))))).... ( -16.90) >DroSec_CAF1 3086 100 - 1 A--AAAACCUUAAGAAAAUAAAAAAAA-GGGGAAAAACUAAAAAUCUACCUGACGCCAUUCG-AUGGCAGUUAAGGCAGGUGUGUGCACUAUUUACACUCGAGA .--....((((..............))-)).............((((.(((((((((((...-))))).))).))).))))(((((.......)))))...... ( -17.54) >DroSim_CAF1 4751 99 - 1 ---AAAACCUUAAGAAAAUAAAAAAAA-G-GAAAAAACUAAAAAUCUACCUGACGCCAUUCGAAUGGCAGUUAAGGCAGGUGUGUGCACUAUUUACACUCGAGA ---....((((..............))-)-)............((((.(((((((((((....))))).))).))).))))(((((.......)))))...... ( -18.24) >DroEre_CAF1 4629 92 - 1 ---AAUACUUAA--------AAAAAAAAAACGAAA-AAUAGGAAUGUACCUGACGCCAUUCGAAUGGCAGUUAAGGCAGGUGUGUGCACUAUUUACACUCGAGA ---.........--------..........(((.(-(((((..(..(((((((((((((....))))).)).....))))))..)...))))))....)))... ( -20.90) >DroYak_CAF1 4784 92 - 1 ---AAUACGUUA--------AAAUAAA-AACGAAAAGCUAGGAAUGUACCUGACGCCAUUCGAAUGGCAGUUAAGGCAGGUGUGUGCACUAUUUACACUCGAGA ---....((((.--------.......-)))).....((..((.(((((((((((((((....))))).))).)))..((((....))))...)))).)).)). ( -19.40) >consensus ___AAUACCUUAAGAAAAUAAAAAAAA_AAGGAAAAACUAAGAAUCUACCUGACGCCAUUCGAAUGGCAGUUAAGGCAGGUGUGUGCACUAUUUACACUCGAGA ...........................................((((.(((((((((((....))))).))).))).))))(((((.......)))))...... (-14.82 = -15.22 + 0.40)


| Location | 13,739,287 – 13,739,401 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -13.70 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13739287 114 + 22407834 CUUAACUGCCAUUCGAAUGGCGUCAGGUAGAUCCUUAGUUUUUCCUU-UUUUUUUUAUUUUCUUAACGUAUUUUUUUUUAGCCAUUUUUGGCGGCAUGCACCUGGCAUUAUUUGC .......(((((....)))))(((((((..((((...(((.......-................))).............((((....)))))).))..)))))))......... ( -24.40) >DroSec_CAF1 3115 98 + 1 CUUAACUGCCAU-CGAAUGGCGUCAGGUAGAUUUUUAGUUUUUCCCC-UUUUUUUUAUUUUCUUAAGGUUUU--UUUUUA-------------GCAUGCACCUGGCAUUAUUUGC .......(((((-...)))))(((((((.(....((((.......((-((..............))))....--...)))-------------)....))))))))......... ( -17.38) >DroSim_CAF1 4780 97 + 1 CUUAACUGCCAUUCGAAUGGCGUCAGGUAGAUUUUUAGUUUUUUC-C-UUUUUUUUAUUUUCUUAAGGUUUU---UUUUA-------------GCAUGCACCUGGCAUUAUUUGC .......(((((....)))))(((((((.(....((((......(-(-((..............))))....---..)))-------------)....))))))))......... ( -19.14) >DroEre_CAF1 4658 101 + 1 CUUAACUGCCAUUCGAAUGGCGUCAGGUACAUUCCUAUU-UUUCGUUUUUUUUUU--------UUAAGUAUU-----UUACCUAUUUUUGGCGGCAUGCACCUGGCAUUAUUUGC .......(((((....)))))(((((((.(((.((((((-(..............--------..)))))..-----...((.......)).)).))).)))))))......... ( -21.69) >DroYak_CAF1 4813 101 + 1 CUUAACUGCCAUUCGAAUGGCGUCAGGUACAUUCCUAGCUUUUCGUU-UUUAUUU--------UAACGUAUU-----UUACCUAUUUUUGGCGGCAUGCACCUGGCAUUAUUUGC .......(((((....)))))(((((((.(((.((........((((-.......--------.))))....-----...((.......)).)).))).)))))))......... ( -24.90) >consensus CUUAACUGCCAUUCGAAUGGCGUCAGGUAGAUUCUUAGUUUUUCCUU_UUUUUUUUAUUUUCUUAACGUAUU___UUUUA_C_AUUUUUGGCGGCAUGCACCUGGCAUUAUUUGC .......(((((....)))))(((((((.....................................................(((....)))..(....))))))))......... (-13.70 = -13.92 + 0.22)

| Location | 13,739,287 – 13,739,401 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -11.81 |
| Energy contribution | -11.81 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
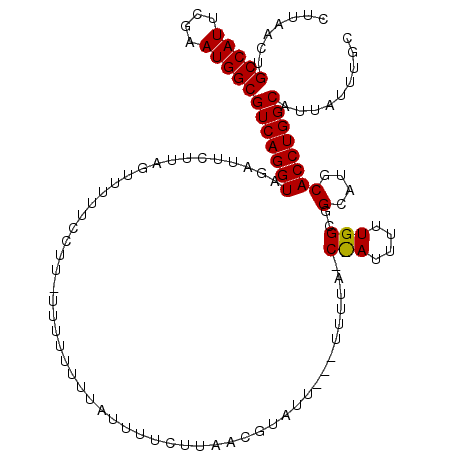
>2L_DroMel_CAF1 13739287 114 - 22407834 GCAAAUAAUGCCAGGUGCAUGCCGCCAAAAAUGGCUAAAAAAAAAUACGUUAAGAAAAUAAAAAAAA-AAGGAAAAACUAAGGAUCUACCUGACGCCAUUCGAAUGGCAGUUAAG (((.....)))((((((.((.((((((....))))................................-.((......))..)))).))))))..(((((....)))))....... ( -22.90) >DroSec_CAF1 3115 98 - 1 GCAAAUAAUGCCAGGUGCAUGC-------------UAAAAA--AAAACCUUAAGAAAAUAAAAAAAA-GGGGAAAAACUAAAAAUCUACCUGACGCCAUUCG-AUGGCAGUUAAG (((.....)))((((((.((..-------------......--....((((...............)-)))............)).))))))..(((((...-)))))....... ( -15.81) >DroSim_CAF1 4780 97 - 1 GCAAAUAAUGCCAGGUGCAUGC-------------UAAAA---AAAACCUUAAGAAAAUAAAAAAAA-G-GAAAAAACUAAAAAUCUACCUGACGCCAUUCGAAUGGCAGUUAAG (((.....)))((((((.((..-------------.....---....((((..............))-)-)............)).))))))..(((((....)))))....... ( -15.54) >DroEre_CAF1 4658 101 - 1 GCAAAUAAUGCCAGGUGCAUGCCGCCAAAAAUAGGUAA-----AAUACUUAA--------AAAAAAAAAACGAAA-AAUAGGAAUGUACCUGACGCCAUUCGAAUGGCAGUUAAG (((.....)))(((((((((.(((((.......)))..-----.....((..--------....)).........-....)).)))))))))..(((((....)))))....... ( -23.20) >DroYak_CAF1 4813 101 - 1 GCAAAUAAUGCCAGGUGCAUGCCGCCAAAAAUAGGUAA-----AAUACGUUA--------AAAUAAA-AACGAAAAGCUAGGAAUGUACCUGACGCCAUUCGAAUGGCAGUUAAG (((.....)))(((((((((.(((((.......)))..-----....((((.--------.......-))))........)).)))))))))..(((((....)))))....... ( -28.80) >consensus GCAAAUAAUGCCAGGUGCAUGCCGCCAAAAAU_G_UAAAA___AAUACCUUAAGAAAAUAAAAAAAA_AAGGAAAAACUAAGAAUCUACCUGACGCCAUUCGAAUGGCAGUUAAG (((.....)))((((((.((...............................................................)).))))))..(((((....)))))....... (-11.81 = -11.81 + -0.00)

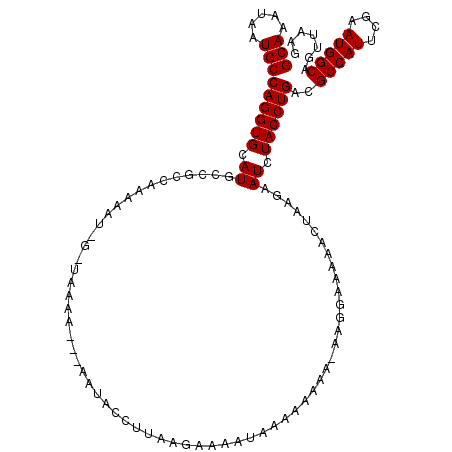
| Location | 13,739,361 – 13,739,461 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.64 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -23.28 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13739361 100 - 22407834 UGGUAAGCACUUGAAGGACGGGCGCCUUAACUCAGUUCUGUUUUUCAUCUUUUAUCUUAUGCAAAUAAUGCCAGGUGCAUGCCGCCAAAAAUGGCUAAAA .((((.(((((((((((((((((...........).))))))))))..............(((.....))).)))))).))))((((....))))..... ( -28.60) >DroSec_CAF1 3186 87 - 1 UGGUAAGCACUUGAAGGACGGGCGCCUUAACUCAGGUCUGUUUUUCAUCUUUUAUCUUAUGCAAAUAAUGCCAGGUGCAUGC-------------UAAAA (((((.(((((((((((((((..((((......)))))))))))))..............(((.....))).)))))).)))-------------))... ( -25.50) >DroSim_CAF1 4850 87 - 1 UGGUAAGCACUUGAAGGACGGGCGCCUUAACUCAGGUCUGUUUUUCAUCUUUUAUCUUAUGCAAAUAAUGCCAGGUGCAUGC-------------UAAAA (((((.(((((((((((((((..((((......)))))))))))))..............(((.....))).)))))).)))-------------))... ( -25.50) >DroEre_CAF1 4721 98 - 1 UGGUAAGCACUUGAAGGACGGGCGCCUUAACUCAGGUCUGUUUUUCAUCUUUUAUCUUAUGCAAAUAAUGCCAGGUGCAUGCCGCCAAAAAUAGGUAA-- .((((.(((((((((((((((..((((......)))))))))))))..............(((.....))).)))))).))))(((.......)))..-- ( -29.00) >DroYak_CAF1 4876 98 - 1 UGGUAAGCACUUGAAGGACGGGCGCCUUAACUUAGGUCUGUUUUUCAUCUUUUAUCUUAUGCAAAUAAUGCCAGGUGCAUGCCGCCAAAAAUAGGUAA-- .((((.(((((((((((((((..((((......)))))))))))))..............(((.....))).)))))).))))(((.......)))..-- ( -29.70) >consensus UGGUAAGCACUUGAAGGACGGGCGCCUUAACUCAGGUCUGUUUUUCAUCUUUUAUCUUAUGCAAAUAAUGCCAGGUGCAUGCCGCCAAAAAU_G_UAAAA .((((.(((((((((((((((..((((......)))))))))))))..............(((.....))).)))))).))))................. (-23.28 = -23.88 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:11 2006