
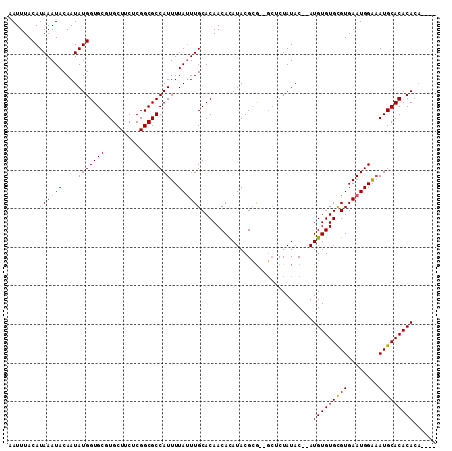
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,738,960 – 13,739,160 |
| Length | 200 |
| Max. P | 0.990432 |

| Location | 13,738,960 – 13,739,069 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.69 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -22.31 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13738960 109 + 22407834 AAUUUACAUAAAUACAAUAUGGUGCGUGCUUCCCGGCGCCAUUUUAUUUGCACAACACAUACGCA--GCUCUAUACAUAUGUGUGUGUGAAUGGAAAUGCACACACACACA .................(((((.(((((((....))))).........(((...........)))--)).)))))....((((((((((.((....)).)))))))))).. ( -29.60) >DroSec_CAF1 2799 103 + 1 AAUUUACAUAAAUACAAUAUGGUGCGUGCUUCUCGGCGCCAUUUUAUUUGCACAACACAUACGCGCGGCUCUAUAC--AUGUGUGCGUGAAUGGGCAUGCACACA------ .................(((((.(((((((....))))).........(((.(.........).))))).))))).--.((((((((((......))))))))))------ ( -27.10) >DroSim_CAF1 4456 107 + 1 AAUUUACAUAAAUACAAUAUGGUGCGUGCUUCUCGGCGCCAUUUUAUUUGCACAACACAUACGCGGUGCUCUAUAC--AUGUGUGCGUGAAUGGAAAUGCACACACAUA-- .........(((((....(((((((..........)))))))..)))))((((..(......)..)))).......--.(((((((((........)))))))))....-- ( -28.50) >DroEre_CAF1 4338 103 + 1 AAUUUACAUAAAUACAAUAUGGUGCGUGCUUCUCGGCGCCAUUUUACUUGCACAACAUAUACGGG--UGUCUAUAC--AUGUGUGCGUGAAUGGAAAUGCACACACA---- ......((((.......))))(((((((((....))).((((((....((((((.(((.((..(.--...)..)).--))))))))).))))))..)))))).....---- ( -26.10) >consensus AAUUUACAUAAAUACAAUAUGGUGCGUGCUUCUCGGCGCCAUUUUAUUUGCACAACACAUACGCG__GCUCUAUAC__AUGUGUGCGUGAAUGGAAAUGCACACACA____ ........((((((....(((((((..........)))))))..)))))).............................(((((((((........)))))))))...... (-22.31 = -22.38 + 0.06)


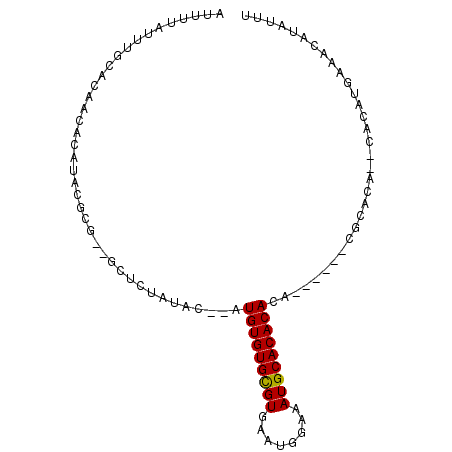
| Location | 13,739,000 – 13,739,093 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -19.88 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.18 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13739000 93 + 22407834 AUUUUAUUUGCACAACACAUACGCA--GCUCUAUACAUAUGUGUGUGUGAAUGGAAAUGCACACACACACA--CGCACACACCCAUGAAACAUAUUU .((((((((((...........)))--)...........((((((((((.((....)).))))))))))..--...........))))))....... ( -18.90) >DroSec_CAF1 2839 81 + 1 AUUUUAUUUGCACAACACAUACGCGCGGCUCUAUAC--AUGUGUGCGUGAAUGGGCAUGCACACA------------CA--CACAUGGAACAUAUUU .......((((.(.........).)))).(((((..--.((((((((((......))))))))))------------..--...)))))........ ( -18.20) >DroSim_CAF1 4496 89 + 1 AUUUUAUUUGCACAACACAUACGCGGUGCUCUAUAC--AUGUGUGCGUGAAUGGAAAUGCACACACAUA----CGCACA--CACAUGGAACAUAUUU .........((((..(......)..))))(((((..--.((((((((((..((..........))..))----))))))--)).)))))........ ( -23.80) >DroEre_CAF1 4378 87 + 1 AUUUUACUUGCACAACAUAUACGGG--UGUCUAUAC--AUGUGUGCGUGAAUGGAAAUGCACACACA----CACACGCA--CAAGCGAAACAUAUUU .......((((....(......).(--(((......--.(((((((((........)))))))))..----.....)))--)..))))......... ( -18.64) >consensus AUUUUAUUUGCACAACACAUACGCG__GCUCUAUAC__AUGUGUGCGUGAAUGGAAAUGCACACACA______CGCACA__CACAUGAAACAUAUUU .......................................(((((((((........)))))))))................................ (-12.36 = -12.18 + -0.19)


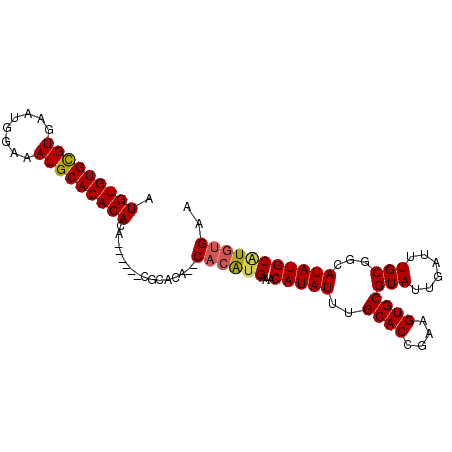
| Location | 13,739,036 – 13,739,133 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -23.40 |
| Energy contribution | -23.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13739036 97 + 22407834 AUGUGUGUGUGAAUGGAAAUGCACACACACACA--CGCACACACCCAUGAAACAUAUUUGCACCAAAGUGCGUAUUGAUUUGCGGCAUAUGCGUGUGAA ..(((((((((.((....)).)))))))))(((--((((.....((..(((.((....(((((....)))))...)).)))..))....)))))))... ( -35.00) >DroSec_CAF1 2875 85 + 1 AUGUGUGCGUGAAUGGGCAUGCACACA------------CA--CACAUGGAACAUAUUUGCACCGAAGUGCGUAUUGAUUUGCGGCAUAUGCAUGUGAA .((((((((((......))))))))))------------..--((((((...(((((..((((....))))(((......)))...))))))))))).. ( -30.10) >DroSim_CAF1 4532 93 + 1 AUGUGUGCGUGAAUGGAAAUGCACACACAUA----CGCACA--CACAUGGAACAUAUUUGCACCGAAGUGCGUAUUGAUUUGCGGCAUAUGCAUGUGAA .(((((((((........))))))))).(((----(((((.--....(((..((....))..)))..))))))))...((..((((....)).))..)) ( -28.20) >DroEre_CAF1 4412 93 + 1 AUGUGUGCGUGAAUGGAAAUGCACACACA----CACACGCA--CAAGCGAAACAUAUUUGCACCGAAGUGCGUAUUGAUUUGCGGCAUAUGCAUGUGAA .(((((((((........)))))))))..----((((.(((--...(((((.((....(((((....)))))...)).)))))......))).)))).. ( -31.70) >consensus AUGUGUGCGUGAAUGGAAAUGCACACACA______CGCACA__CACAUGAAACAUAUUUGCACCGAAGUGCGUAUUGAUUUGCGGCAUAUGCAUGUGAA .(((((((((........)))))))))................((((((...(((((..((((....))))(((......)))...))))))))))).. (-23.40 = -23.65 + 0.25)

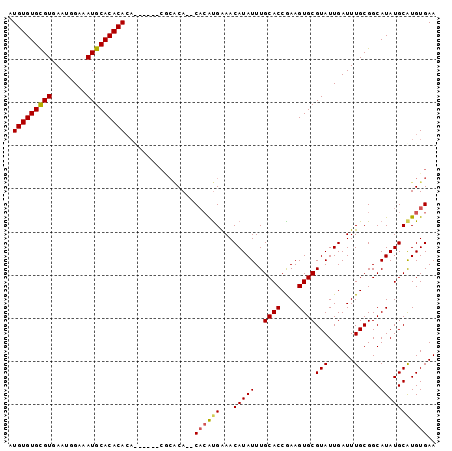
| Location | 13,739,036 – 13,739,133 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.93 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -4.76 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13739036 97 - 22407834 UUCACACGCAUAUGCCGCAAAUCAAUACGCACUUUGGUGCAAAUAUGUUUCAUGGGUGUGUGCG--UGUGUGUGUGUGCAUUUCCAUUCACACACACAU ...(((((((((((((............((((....))))...((((...))))))))))))))--)))(((((((((.((....)).))))))))).. ( -39.70) >DroSec_CAF1 2875 85 - 1 UUCACAUGCAUAUGCCGCAAAUCAAUACGCACUUCGGUGCAAAUAUGUUCCAUGUG--UG------------UGUGUGCAUGCCCAUUCACGCACACAU ...((((((.......))..........((((....))))....)))).....(((--((------------((((((.((....)).))))))))))) ( -26.30) >DroSim_CAF1 4532 93 - 1 UUCACAUGCAUAUGCCGCAAAUCAAUACGCACUUCGGUGCAAAUAUGUUCCAUGUG--UGUGCG----UAUGUGUGUGCAUUUCCAUUCACGCACACAU .......(((((((((((..........((((....))))..(((((...))))))--)).)))----)))))((((((............)))))).. ( -31.90) >DroEre_CAF1 4412 93 - 1 UUCACAUGCAUAUGCCGCAAAUCAAUACGCACUUCGGUGCAAAUAUGUUUCGCUUG--UGCGUGUG----UGUGUGUGCAUUUCCAUUCACGCACACAU ...(((((((((.((.(.((((......((((....))))......))))))).))--)))))))(----((((((((.((....)).))))))))).. ( -34.70) >consensus UUCACAUGCAUAUGCCGCAAAUCAAUACGCACUUCGGUGCAAAUAUGUUCCAUGUG__UGUGCG______UGUGUGUGCAUUUCCAUUCACGCACACAU ...((((((.......))..........((((....))))....))))......................((((((((.((....)).))))))))... (-17.36 = -17.93 + 0.56)

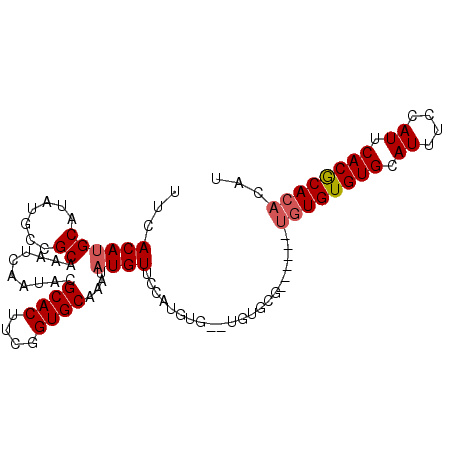
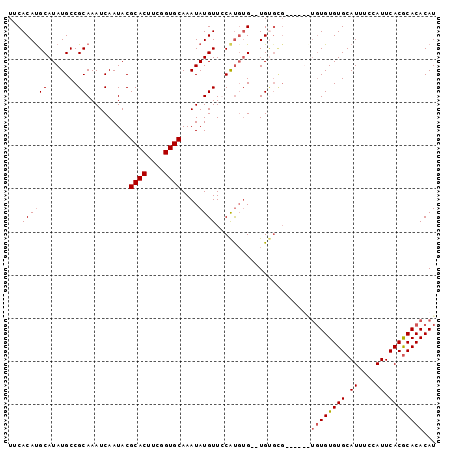
| Location | 13,739,069 – 13,739,160 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 89.11 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13739069 91 - 22407834 UUU-UUUGUAUCGAAACGAGGAAAGCAUUUCACACGCAUAUGCCGCAAAUCAAUACGCACUUUGGUGCAAAUAUGUUUCAUGGGUGUGUGCG-- (((-(((((......))))))))...........((((((((((............((((....))))...((((...))))))))))))))-- ( -23.40) >DroSec_CAF1 2902 86 - 1 UUUUUUUGUAUCGGAACGAGGAAAGCAUUUCACAUGCAUAUGCCGCAAAUCAAUACGCACUUCGGUGCAAAUAUGUUCCAUGUG--UG------ ............((((((.((...((((.....)))).....))............((((....)))).....)))))).....--..------ ( -21.20) >DroSim_CAF1 4563 90 - 1 UUUUUUUGUAUCGGAACGAGGAAAGCAUUUCACAUGCAUAUGCCGCAAAUCAAUACGCACUUCGGUGCAAAUAUGUUCCAUGUG--UGUGCG-- (((((((((......)))))))))((((..(((((((((((...(.....).....((((....))))..)))))...))))))--.)))).-- ( -25.00) >DroEre_CAF1 4441 91 - 1 UUU-UUUGUAUCGGAACGAGGAAAACAUUUCACAUGCAUAUGCCGCAAAUCAAUACGCACUUCGGUGCAAAUAUGUUUCGCUUG--UGCGUGUG (((-(((((......)))))))).......((((((((((.((.(.((((......((((....))))......))))))).))--)))))))) ( -27.00) >consensus UUU_UUUGUAUCGGAACGAGGAAAGCAUUUCACAUGCAUAUGCCGCAAAUCAAUACGCACUUCGGUGCAAAUAUGUUCCAUGUG__UGUGCG__ ............((((((.((...((((.....)))).....))............((((....)))).....))))))............... (-17.19 = -17.25 + 0.06)

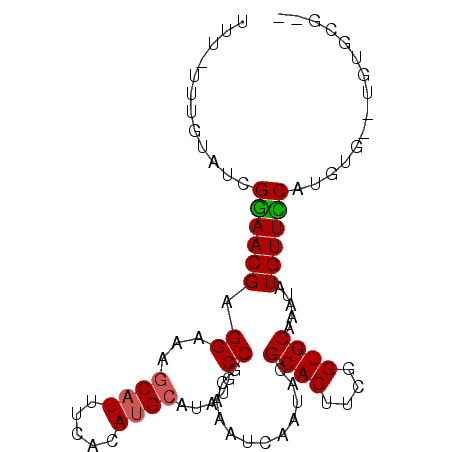
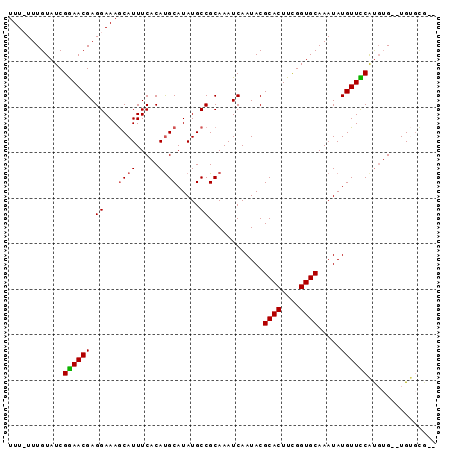
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:07 2006