
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,738,712 – 13,738,817 |
| Length | 105 |
| Max. P | 0.658087 |

| Location | 13,738,712 – 13,738,817 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -13.42 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13738712 105 + 22407834 GAACAAGAUGAAAACAAAAAUCUGUUAUUCAGAUUGACUUGGC--------------ACA-AAAGCCGACCUCGGCCAAAAAGCUUGUACAUAUAUAUGUAUGUAUUCAGUUUUUCCUUG ...((((.......(((.((((((.....))))))...)))..--------------...-...((((....))))..(((((((.((((((((....))))))))..))))))).)))) ( -25.20) >DroSec_CAF1 2551 105 + 1 GAACAAGAUGAAAACAAAAAUCUGUUAUUCAGAUUGACUUGGC--------------ACA-AAAGCCGACCUCGGCCAAAAAGCUUGUACAUAUAUAUGUAUGUAUUCAGUUUUUCUUUG ...(((..((((((((......)))).))))..))).......--------------.((-((.((((....))))..(((((((.((((((((....))))))))..))))))).)))) ( -24.20) >DroAna_CAF1 14546 111 + 1 CAAGAUGAUGGCAACA-AAAUCUGUUAUUCAGAUUGACUUGCCACAUUUAACAUGCCACA-AAGGCCUACCUCGGCG-----CCUCGAACGAAAACGCGUA--UUUGCAGUUUCUCCUUU ..(((((.((((((..-.((((((.....))))))...))))))))))).....(((...-.(((....))).))).-----........((((..(((..--..)))..))))...... ( -28.20) >DroSim_CAF1 4208 105 + 1 GAACAAGAUGAAAACAAAAAUCUGUUAUUCAGAUUGACUUGGC--------------ACA-AAAGCCGACCUCGGCCAAAAAGCUUGUACAUAUAUAUGUAUGUAUUCAGUUUUUCUUUG ...(((..((((((((......)))).))))..))).......--------------.((-((.((((....))))..(((((((.((((((((....))))))))..))))))).)))) ( -24.20) >DroEre_CAF1 4095 103 + 1 GAACAAGAUGAAAACAAAAAUCUGUUAUUCAGAUUGACUUGGC--------------ACA-AAAGCCGACCUCGGCCAAAAAGCAUGUAUAUAUACA--UAUAUAUUCAGUUUUUCUUUG .....(((.((((((...((((((.....))))))((....((--------------...-...((((....))))......))((((((((....)--))))))))).)))))).))). ( -19.80) >DroYak_CAF1 4246 96 + 1 GAACAAGAUGAAAACCAAAAUCUGUUAUUCAGAUUGACUUGGC--------------ACACAAAGGCGACCUCAGCCAAAUAGCGUGUACACG----------AUUUCAGUUUUUCUUUG (((.(((.(((((.((((((((((.....))))))...)))).--------------.......(((.......)))......(((....)))----------.))))).)))))).... ( -17.20) >consensus GAACAAGAUGAAAACAAAAAUCUGUUAUUCAGAUUGACUUGGC______________ACA_AAAGCCGACCUCGGCCAAAAAGCUUGUACAUAUAUAUGUAUGUAUUCAGUUUUUCUUUG ...((((..((((((...((((((.....))))))....(((......................((((....))))..........(((((((......)))))))))))))))).)))) (-13.42 = -14.32 + 0.89)

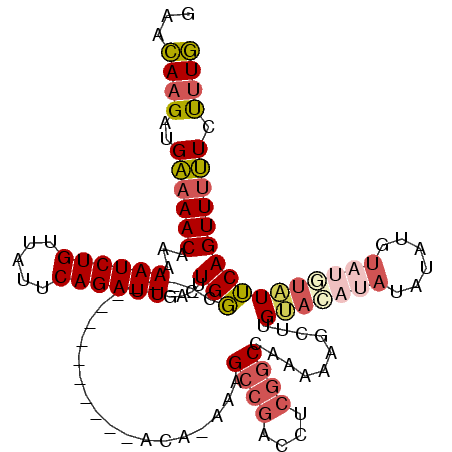
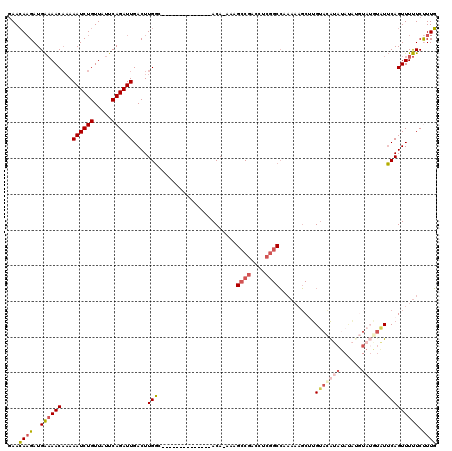
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:03 2006