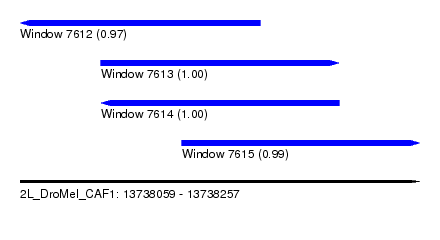
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,738,059 – 13,738,257 |
| Length | 198 |
| Max. P | 0.999908 |

| Location | 13,738,059 – 13,738,178 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -30.42 |
| Energy contribution | -31.29 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13738059 119 - 22407834 UGGCUCUU-UGGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAAUCAACCCAGACAAAGGCCAAACUUUUUGUCUACAUCUCGGUGUCGCUUAGUGAAUGCCA .(((...(-(((((((((((.........((((((.((((((....))))))))))))...........))))))))))))(((....((..((((....)))).))..)))....))). ( -37.45) >DroSec_CAF1 1899 119 - 1 UGGCUCUU-UGGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAAUCAACUCAGACAAAGGCCAAACUUUUUGUCUACAUCUCGGUGUCGCUUAGUGAAUGCCA .(((...(-(((((((((((.........((((((.((((((....))))))))))))...........))))))))))))(((....((..((((....)))).))..)))....))). ( -37.45) >DroAna_CAF1 13921 119 - 1 UGGCUCUUUUGGCUUUUGUUGCUGCUAAAAGCCAAACAAUAGCCGUCUAUUGUUUGCCGAACAAACUCAGACAAAGGCCAAACUU-UUGUCUCCAUCUCGCAGUCGCUUAGUGAAUUCCA .(((......(((((((((....)).)))))))(((((((((....))))))))))))(((...(((.(((((((((.....)))-)))))).......((....))..)))...))).. ( -32.20) >DroSim_CAF1 3548 119 - 1 UGGCUCUU-UGGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAAUCAACUCAGACAAAGGCCAAACUUUUUGUCUACAUCUCGGUGUCGCUUAGUGAAUGCCA .(((...(-(((((((((((.........((((((.((((((....))))))))))))...........))))))))))))(((....((..((((....)))).))..)))....))). ( -37.45) >DroEre_CAF1 3426 119 - 1 UGGCUCUU-ACGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAACCAACGCAGACAAAGGCCAAACUUUUUGUCUACAACUCGGUGUCGCCUAGUGAAUUCCA (((((.((-(.((..........))))).)))))..((..(((((((....(((((.....)))))(.(((((((((......))))))))).).....))))..)))...))....... ( -32.50) >DroYak_CAF1 3359 119 - 1 UGGCUCUU-UGGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAAUCAACUCAGACAAAGGCCAAACUUUUUGUCUACAUCUCGGUGUCGCUUAGUGAAUUCCA .(((..((-(((((((((((.........((((((.((((((....))))))))))))...........)))))))))))))......)))........((..((((...))))...)). ( -35.35) >consensus UGGCUCUU_UGGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAAUCAACUCAGACAAAGGCCAAACUUUUUGUCUACAUCUCGGUGUCGCUUAGUGAAUGCCA .(((.....(((((((((((.........((((((.((((((....))))))))))))...........)))))))))))........)))........((((((((...))).))))). (-30.42 = -31.29 + 0.86)


| Location | 13,738,099 – 13,738,217 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.96 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -33.10 |
| Energy contribution | -33.38 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13738099 118 + 22407834 UGGCCUUUGUCUGGGUUGAUUCAGCCAACAAUAGGCGGCCAUUGUUUGGCUUUUAGCAGCAACAAAAGCCA-AAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCA-ACGGAAAUUAAAU ((((..((((((...((((....(((.......)))....((((((((((((((.((..........))..-))))))))))))))))))...))))))...))))-............. ( -35.30) >DroSec_CAF1 1939 118 + 1 UGGCCUUUGUCUGAGUUGAUUCAGCCAACAAUAGGCGGCCAUUGUUUGGCUUUUAGCAGCAACAAAAGCCA-AAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCA-ACGGAAAUUAAAU ((((..((((((...((((....(((.......)))....((((((((((((((.((..........))..-))))))))))))))))))...))))))...))))-............. ( -35.30) >DroAna_CAF1 13960 120 + 1 UGGCCUUUGUCUGAGUUUGUUCGGCAAACAAUAGACGGCUAUUGUUUGGCUUUUAGCAGCAACAAAAGCCAAAAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCAGAAGGAAAUUAAAU ...((((((((((((((.(((.((((((((((((....)))))))))(((((((.(......))))))))......))).)))))))))))).((........)).)))))......... ( -36.40) >DroSim_CAF1 3588 118 + 1 UGGCCUUUGUCUGAGUUGAUUCAGCCAACAAUAGGCGGCCAUUGUUUGGCUUUUAGCAGCAACAAAAGCCA-AAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCA-ACGGAAAUUAAAU ((((..((((((...((((....(((.......)))....((((((((((((((.((..........))..-))))))))))))))))))...))))))...))))-............. ( -35.30) >DroEre_CAF1 3466 118 + 1 UGGCCUUUGUCUGCGUUGGUUCAGCCAACAAUAGGCGGCCAUUGUUUGGCUUUUAGCAGCAACAAAAGCGU-AAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCA-ACGGAAAUUAAAU ((((..((((((..((((((...(((.......))).)))(((((((((((((((((..........)).)-))))))))))))))...))).))))))...))))-............. ( -40.20) >DroYak_CAF1 3399 118 + 1 UGGCCUUUGUCUGAGUUGAUUCAGCCAACAAUAGGCGGCCAUUGUUUGGCUUUUAGCAGCAACAAAAGCCA-AAGAGCCAAACAAUUUAGAUGGGACAAUAAGCCA-ACGGAAAUUAAAU ((((..((((((.(.((((....(((.......)))....((((((((((((((.((..........))..-)))))))))))))))))).).))))))...))))-............. ( -35.20) >consensus UGGCCUUUGUCUGAGUUGAUUCAGCCAACAAUAGGCGGCCAUUGUUUGGCUUUUAGCAGCAACAAAAGCCA_AAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCA_ACGGAAAUUAAAU ((((..((((((...((((....(((.......)))....((((((((((((((....((.......))...))))))))))))))))))...))))))...)))).............. (-33.10 = -33.38 + 0.28)


| Location | 13,738,099 – 13,738,217 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.96 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -33.42 |
| Energy contribution | -33.92 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.28 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.07 |
| SVM RNA-class probability | 0.999785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13738099 118 - 22407834 AUUUAAUUUCCGU-UGGCUUAUUGUCCUAUCUAAAUUGUUUGGCUCUU-UGGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAAUCAACCCAGACAAAGGCCA .............-((((((.(((((........(((((((((((.((-((((..........)))))))))))))))))(((((.(....).)))))...........))))))))))) ( -38.60) >DroSec_CAF1 1939 118 - 1 AUUUAAUUUCCGU-UGGCUUAUUGUCCUAUCUAAAUUGUUUGGCUCUU-UGGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAAUCAACUCAGACAAAGGCCA .............-((((((.(((((........(((((((((((.((-((((..........)))))))))))))))))(((((.(....).)))))...........))))))))))) ( -38.60) >DroAna_CAF1 13960 120 - 1 AUUUAAUUUCCUUCUGGCUUAUUGUCCUAUCUAAAUUGUUUGGCUCUUUUGGCUUUUGUUGCUGCUAAAAGCCAAACAAUAGCCGUCUAUUGUUUGCCGAACAAACUCAGACAAAGGCCA ...............(((((.(((((.........(((((((((......(((((((((....)).)))))))(((((((((....)))))))))))))))))).....)))))))))). ( -38.44) >DroSim_CAF1 3588 118 - 1 AUUUAAUUUCCGU-UGGCUUAUUGUCCUAUCUAAAUUGUUUGGCUCUU-UGGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAAUCAACUCAGACAAAGGCCA .............-((((((.(((((........(((((((((((.((-((((..........)))))))))))))))))(((((.(....).)))))...........))))))))))) ( -38.60) >DroEre_CAF1 3466 118 - 1 AUUUAAUUUCCGU-UGGCUUAUUGUCCUAUCUAAAUUGUUUGGCUCUU-ACGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAACCAACGCAGACAAAGGCCA .............-((((((.(((((........(((((((((((.((-(.((..........))))).)))))))))))(((((.(....).)))))...........))))))))))) ( -35.40) >DroYak_CAF1 3399 118 - 1 AUUUAAUUUCCGU-UGGCUUAUUGUCCCAUCUAAAUUGUUUGGCUCUU-UGGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAAUCAACUCAGACAAAGGCCA .............-((((((.(((((........(((((((((((.((-((((..........)))))))))))))))))(((((.(....).)))))...........))))))))))) ( -38.60) >consensus AUUUAAUUUCCGU_UGGCUUAUUGUCCUAUCUAAAUUGUUUGGCUCUU_UGGCUUUUGUUGCUGCUAAAAGCCAAACAAUGGCCGCCUAUUGUUGGCUGAAUCAACUCAGACAAAGGCCA ...............(((((.(((((........(((((((((((.((..(((.......)))...)).)))))))))))(((((.(....).)))))...........)))))))))). (-33.42 = -33.92 + 0.50)

| Location | 13,738,139 – 13,738,257 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -25.41 |
| Energy contribution | -25.44 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13738139 118 + 22407834 AUUGUUUGGCUUUUAGCAGCAACAAAAGCCA-AAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCA-ACGGAAAUUAAAUUGGCUGCUCUAGUUUACAUAGUAGUUCUCGCCCAUCAUCU ((((((((((((((.((..........))..-))))))))))))))...(((.((.(....(((((-(...........))))))((((((.......))).)))....))).))).... ( -28.90) >DroSec_CAF1 1979 118 + 1 AUUGUUUGGCUUUUAGCAGCAACAAAAGCCA-AAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCA-ACGGAAAUUAAAUUGGCUGCUCUAGUUUACAUAGUAGUUCUCGCCCAUCAUCA ((((((((((((((.((..........))..-))))))))))))))...(((.((.(....(((((-(...........))))))((((((.......))).)))....))).))).... ( -28.90) >DroAna_CAF1 14000 108 + 1 AUUGUUUGGCUUUUAGCAGCAACAAAAGCCAAAAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCAGAAGGAAAUUAAAUUGGCUGCUCCACUUUACAAACUACAC-----------UC- ((((((((((((((....((.......))...)))))))))))))).......(((.....((((((............))))))..)))................-----------..- ( -25.20) >DroSim_CAF1 3628 118 + 1 AUUGUUUGGCUUUUAGCAGCAACAAAAGCCA-AAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCA-ACGGAAAUUAAAUUGGCUGCUCUAGUUUACAUAGUAGUUCUCGCCCAUCAUCA ((((((((((((((.((..........))..-))))))))))))))...(((.((.(....(((((-(...........))))))((((((.......))).)))....))).))).... ( -28.90) >DroEre_CAF1 3506 118 + 1 AUUGUUUGGCUUUUAGCAGCAACAAAAGCGU-AAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCA-ACGGAAAUUAAAUUGGCUGCUCUAGUUUACAUAGUAGUUCUCGCCCAUCAUCU (((((((((((((((((..........)).)-))))))))))))))...(((.((.(....(((((-(...........))))))((((((.......))).)))....))).))).... ( -31.40) >DroYak_CAF1 3439 118 + 1 AUUGUUUGGCUUUUAGCAGCAACAAAAGCCA-AAGAGCCAAACAAUUUAGAUGGGACAAUAAGCCA-ACGGAAAUUAAAUUGGCUGCUCUAGUUUACAUAGUAGUUCUCGCCUAUCAUCU ((((((((((((((.((..........))..-))))))))))))))...((((((.(....(((((-(...........))))))((((((.......))).)))....))))))).... ( -33.10) >consensus AUUGUUUGGCUUUUAGCAGCAACAAAAGCCA_AAGAGCCAAACAAUUUAGAUAGGACAAUAAGCCA_ACGGAAAUUAAAUUGGCUGCUCUAGUUUACAUAGUAGUUCUCGCCCAUCAUCU ((((((((((((((....((.......))...)))))))))))))).(((((.(((.....(((((..............)))))..))).)))))........................ (-25.41 = -25.44 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:02 2006