
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,737,512 – 13,737,629 |
| Length | 117 |
| Max. P | 0.543978 |

| Location | 13,737,512 – 13,737,629 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
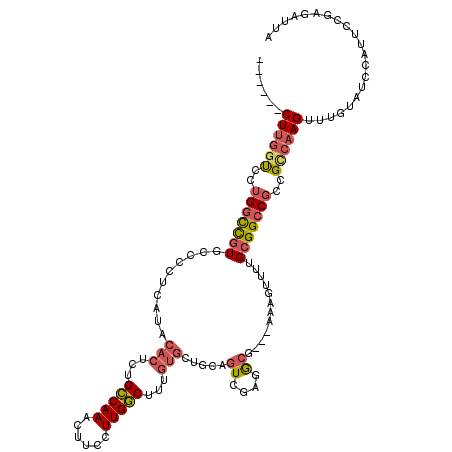
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -22.76 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
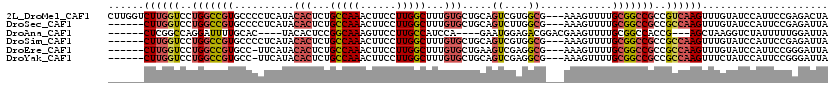
>2L_DroMel_CAF1 13737512 117 + 22407834 CUUGGUCUUGGUCCUGGCCGUGCCCCUCAUACACUCUGCCAAACUUCCUUGGCUUUGUGCUGCAGUCGUGGCG---AAAGUUUUGCGGCCGCCGUCAAGUUUGUAUCCAUUCCGAGACUA ..(((((((((...((((.(((.........)))...)))).((...((((((..((.(((((((....(((.---...))))))))))))..))))))...)).......))))))))) ( -37.70) >DroSec_CAF1 1356 111 + 1 ------CUUGGUCCUGGCCGUGCCCCUCAUACACUCUGCCAAACUUCCUUGGCUUUGUGCUGCAGUCUUGGCG---AAAGUUUUGCGGCCGCCGCCAAGUUUGUAUCCAUUCCGAGAUUA ------(((((...((((.(((.........)))...)))).((...((((((..((.(((((((....(((.---...))))))))))))..))))))...)).......))))).... ( -34.10) >DroAna_CAF1 13388 103 + 1 ------CUCGGCCAGGAUUUUGCAC----UACACUCCGGCAAAGUUCCUUGCCAUCCA----GAAUGGAGACGGACGAAGUUUUGCGGCCACCG---AGCUAAGGUCUAUUUUUGGAUUA ------...(((.((((((((((.(----........))))))).)))).)))(((((----(((.(....)((((..(((....(((...)))---.)))...))))..)))))))).. ( -30.90) >DroSim_CAF1 3005 111 + 1 ------CUUGGUCCUGGCCGUGCCCCUCAUACACUCUGCCAAACUUCCUUGGCUUUGUGCUGCAGUCGUGGCG---AAAGUUUUGCGGCCGCCGCCAAGUUUGUAUCCAUUCCGAGAUUA ------((((((..(((((((((..((((..(((...(((((......)))))...))).)).))..))(((.---...)))..)))))))..))))))..................... ( -34.10) >DroEre_CAF1 2896 110 + 1 ------CUUGGUCCUGGCCGUGCC-UUCAUACACUCUGCCAAACUUCCUUGGCUUUGUGCUGAAGUCGAGGCG---AAAGUUUUGCGGCCGCCGCCAAGUUUGUAUCCAUUCCGGGAUUA ------((((((..((((((((.(-((((..(((...(((((......)))))...))).))))).)(((((.---...))))))))))))..)))))).....((((......)))).. ( -42.70) >DroYak_CAF1 2809 110 + 1 ------CUUGGUCCUGGCCGUGCC-UUCAUACACUCUGCCAAACUUCCUUGGCUUUGUGCUGCAGUCGAGGCG---AAAGUUUUGCGGCCGCCGCCAAGUUUCUAUCCAUUCCGGGAUUA ------((((((..((((((((.(-(.((..(((...(((((......)))))...))).)).)).)(((((.---...))))))))))))..)))))).....((((......)))).. ( -38.30) >consensus ______CUUGGUCCUGGCCGUGCCCCUCAUACACUCUGCCAAACUUCCUUGGCUUUGUGCUGCAGUCGAGGCG___AAAGUUUUGCGGCCGCCGCCAAGUUUGUAUCCAUUCCGAGAUUA ......((((((..(((((((..........(((...(((((......)))))...))).....((....))............)))))))..))))))..................... (-22.76 = -23.48 + 0.73)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:58 2006