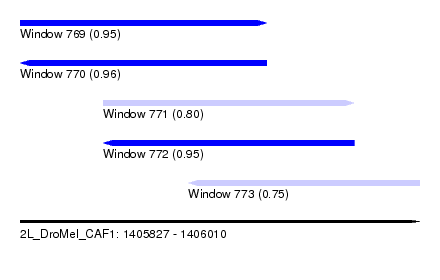
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,405,827 – 1,406,010 |
| Length | 183 |
| Max. P | 0.964915 |

| Location | 1,405,827 – 1,405,940 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.37 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -10.60 |
| Energy contribution | -12.85 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1405827 113 + 22407834 CCAUAAGAACAUAAUUUAAUUAACAAUUUCUAUAUCUUUUAAUGUUGUUCCUCUC-UUAGGAGCUGAGCUCAUAAGGUUUUAAAAUACCCUUCGCUUAGAGUAUCCACUUAGCG ...((((..............((((((................))))))......-...(((.((((((......(((........)))....))))))....))).))))... ( -15.89) >DroSec_CAF1 24063 108 + 1 CCAUAACAAUUUAAUUGAAUUAACAACUUCUAUAUCUUUUAAUGUUGUUCCACUC-CUAGGAG-----AUCAUAUUGUUAAUGAAUACCCUUUGGUUAGGGUAUUCACUUAGCG ...(((((((.....(((...((((((................))))))...(((-....)))-----.))).))))))).((((((((((......))))))))))....... ( -26.49) >DroSim_CAF1 24264 95 + 1 CCAUAACAAUUUAAUUUAAUUAACAAUUUCUAUAUCUUU-------------CUC-CUAGGCG-----AUCAUAUUGUUAAUGAAUACCCUUUGGUUAGGGUAUUCACUUAGCG ..................(((((((((.......(((..-------------...-..)))..-----.....)))))))))(((((((((......)))))))))........ ( -19.64) >DroYak_CAF1 24408 97 + 1 CCACUAUCC---CAUU---------AUUGCUAUAUCUUGUAAUAUUGCUUCACUCCCCAAGAG-----CUCUGAAUGUUUUGAGAUACCCUGCGUUUAGGGUAUCCACUUAGCG .........---((.(---------(((((........)))))).)).(((((((.....)))-----...)))).(((.((.(((((((((....)))))))))))...))). ( -20.90) >consensus CCAUAACAAUUUAAUUUAAUUAACAAUUUCUAUAUCUUUUAAUGUUGUUCCACUC_CUAGGAG_____AUCAUAAUGUUAAUAAAUACCCUUCGGUUAGGGUAUCCACUUAGCG ............................................................................(((((((((((((((......)))))))))).))))). (-10.60 = -12.85 + 2.25)

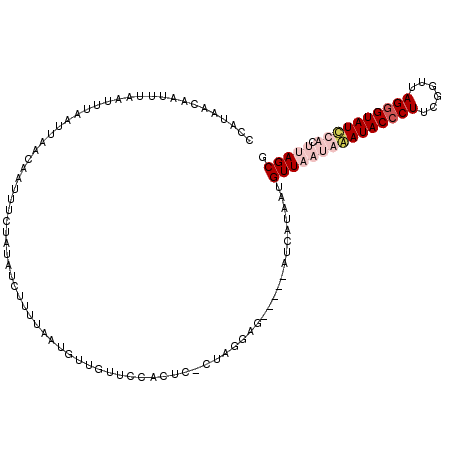
| Location | 1,405,827 – 1,405,940 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.37 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -12.45 |
| Energy contribution | -11.57 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1405827 113 - 22407834 CGCUAAGUGGAUACUCUAAGCGAAGGGUAUUUUAAAACCUUAUGAGCUCAGCUCCUAA-GAGAGGAACAACAUUAAAAGAUAUAGAAAUUGUUAAUUAAAUUAUGUUCUUAUGG ........(((((((((......)))))))))......((((.((((...)))).)))-)..(((((((...((((..((((.......))))..))))....))))))).... ( -19.80) >DroSec_CAF1 24063 108 - 1 CGCUAAGUGAAUACCCUAACCAAAGGGUAUUCAUUAACAAUAUGAU-----CUCCUAG-GAGUGGAACAACAUUAAAAGAUAUAGAAGUUGUUAAUUCAAUUAAAUUGUUAUGG .....((((((((((((......))))))))))))((((((.((((-----(((....-)))...((((((................))))))......)))).)))))).... ( -26.79) >DroSim_CAF1 24264 95 - 1 CGCUAAGUGAAUACCCUAACCAAAGGGUAUUCAUUAACAAUAUGAU-----CGCCUAG-GAG-------------AAAGAUAUAGAAAUUGUUAAUUAAAUUAAAUUGUUAUGG .....((((((((((((......))))))))))))((((((.((((-----(.(....-).)-------------)..((((.......)))).......))).)))))).... ( -20.10) >DroYak_CAF1 24408 97 - 1 CGCUAAGUGGAUACCCUAAACGCAGGGUAUCUCAAAACAUUCAGAG-----CUCUUGGGGAGUGAAGCAAUAUUACAAGAUAUAGCAAU---------AAUG---GGAUAGUGG (((((..((((((((((......)))))))).))...((((....(-----(((.....))))...((.(((((....))))).))...---------))))---...))))). ( -26.00) >consensus CGCUAAGUGAAUACCCUAACCAAAGGGUAUUCAAAAACAAUAUGAG_____CUCCUAG_GAGUGGAACAACAUUAAAAGAUAUAGAAAUUGUUAAUUAAAUUAAAUUGUUAUGG ........(((((((((......))))))))).................................................................................. (-12.45 = -11.57 + -0.87)

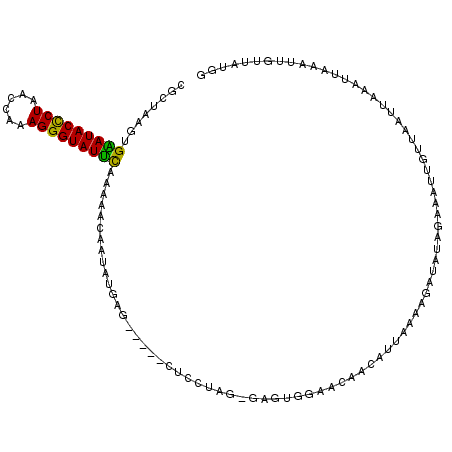
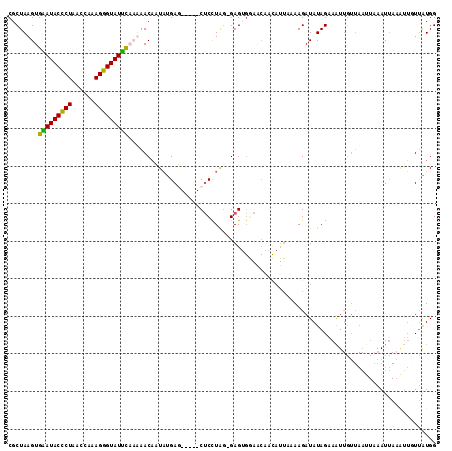
| Location | 1,405,865 – 1,405,980 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.07 |
| Mean single sequence MFE | -27.41 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.88 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1405865 115 + 22407834 UUAAUGUUGUUCCUCUC-UUAGGAGCUGAGCUCAUAAGGUUUUAAAAUACCCUUCGCUUAGAGUAUCCACUUAGCGAUGAAAUAAAAAGCGAAAAAAUAAACAGCCUCAUCGGCCC ....((((((((((...-..))))))...(((.....(((........)))..(((((..((((....)))))))))..........))).........))))(((.....))).. ( -23.20) >DroSec_CAF1 24101 110 + 1 UUAAUGUUGUUCCACUC-CUAGGAG-----AUCAUAUUGUUAAUGAAUACCCUUUGGUUAGGGUAUUCACUUAGCGAUGAAAUAAAAAGCGAAAAAAUAAACAGCCUCAUCGGCCC ....((((.((((.(((-....)))-----....(((((((((((((((((((......)))))))))).))))))))).........).)))......))))(((.....))).. ( -31.00) >DroSim_CAF1 24302 97 + 1 U-------------CUC-CUAGGCG-----AUCAUAUUGUUAAUGAAUACCCUUUGGUUAGGGUAUUCACUUAGCGAUGAAAUAAAAAGCGAAAAAAUAAACAGCCUCAUCGGCCC .-------------...-...((((-----((..(((((((((((((((((((......)))))))))).))))))))).........((.............))...))).))). ( -27.02) >DroEre_CAF1 24067 101 + 1 -----AUUGCUCCACUU--CAGAAG-----CUCGGCGUGUUUA-GGAUACCCUGCGU-UAGGGUAUCCACUUAGCGAUGGAAUAA-AAGCGAAAAAGCAAACAGCCUCAGCGGCCC -----............--.....(-----((.(((.(((((.-((((((((((...-)))))))))).................-..((......))))))))))..)))..... ( -29.50) >DroYak_CAF1 24434 111 + 1 GUAAUAUUGCUUCACUCCCCAAGAG-----CUCUGAAUGUUUUGAGAUACCCUGCGUUUAGGGUAUCCACUUAGCGAUGGAAUAAAAAGCGAAAGAGUAAACAGCCUCAUUGGCCC ......((((((......(((...(-----((..(.......((.(((((((((....))))))))))))..)))..)))......))))))...........(((.....))).. ( -26.31) >consensus UUAAUAUUGCUCCACUC_CUAGGAG_____CUCAUAAUGUUUAUGAAUACCCUUCGGUUAGGGUAUCCACUUAGCGAUGAAAUAAAAAGCGAAAAAAUAAACAGCCUCAUCGGCCC .....................................((((...(((((((((......))))))))).....((.............)).........))))(((.....))).. (-15.20 = -15.88 + 0.68)

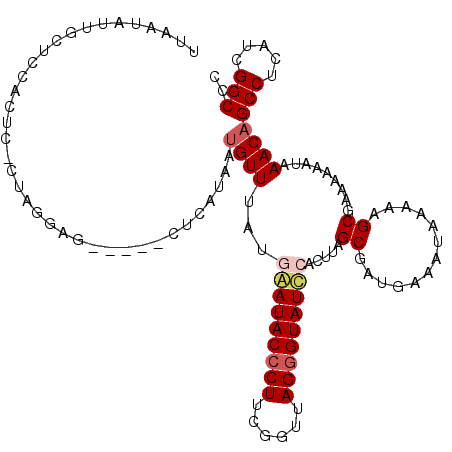
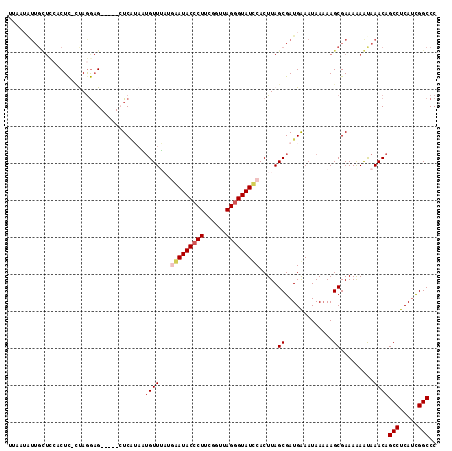
| Location | 1,405,865 – 1,405,980 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.07 |
| Mean single sequence MFE | -30.09 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.74 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1405865 115 - 22407834 GGGCCGAUGAGGCUGUUUAUUUUUUCGCUUUUUAUUUCAUCGCUAAGUGGAUACUCUAAGCGAAGGGUAUUUUAAAACCUUAUGAGCUCAGCUCCUAA-GAGAGGAACAACAUUAA ((((..((((((...((((.((((((((((.....(((((......)))))......)))))))))).....)))).))))))..)))).(.((((..-...)))).)........ ( -28.30) >DroSec_CAF1 24101 110 - 1 GGGCCGAUGAGGCUGUUUAUUUUUUCGCUUUUUAUUUCAUCGCUAAGUGAAUACCCUAACCAAAGGGUAUUCAUUAACAAUAUGAU-----CUCCUAG-GAGUGGAACAACAUUAA .((((.....))))........((((((((((....((((.....((((((((((((......))))))))))))......)))).-----.....))-))))))))......... ( -31.70) >DroSim_CAF1 24302 97 - 1 GGGCCGAUGAGGCUGUUUAUUUUUUCGCUUUUUAUUUCAUCGCUAAGUGAAUACCCUAACCAAAGGGUAUUCAUUAACAAUAUGAU-----CGCCUAG-GAG-------------A (((((((((((((.............))).......))))))...((((((((((((......))))))))))))...........-----.))))..-...-------------. ( -27.43) >DroEre_CAF1 24067 101 - 1 GGGCCGCUGAGGCUGUUUGCUUUUUCGCUU-UUAUUCCAUCGCUAAGUGGAUACCCUA-ACGCAGGGUAUCC-UAAACACGCCGAG-----CUUCUG--AAGUGGAGCAAU----- .((((.....))))..((((((...(((((-(.......((((.....(((((((((.-....)))))))))-.......).))).-----.....)--))))))))))).----- ( -32.82) >DroYak_CAF1 24434 111 - 1 GGGCCAAUGAGGCUGUUUACUCUUUCGCUUUUUAUUCCAUCGCUAAGUGGAUACCCUAAACGCAGGGUAUCUCAAAACAUUCAGAG-----CUCUUGGGGAGUGAAGCAAUAUUAC .((((.....))))............(((((...(((((..(((.((((((((((((......))))))))......))))...))-----)...)))))...)))))........ ( -30.20) >consensus GGGCCGAUGAGGCUGUUUAUUUUUUCGCUUUUUAUUUCAUCGCUAAGUGGAUACCCUAAACGAAGGGUAUUCAUAAACAAUAUGAG_____CUCCUAG_GAGUGGAACAACAUUAA .((((.....))))........((((((((.(((..............(((((((((......)))))))))...........(((.....))).))).))))))))......... (-20.58 = -20.74 + 0.16)

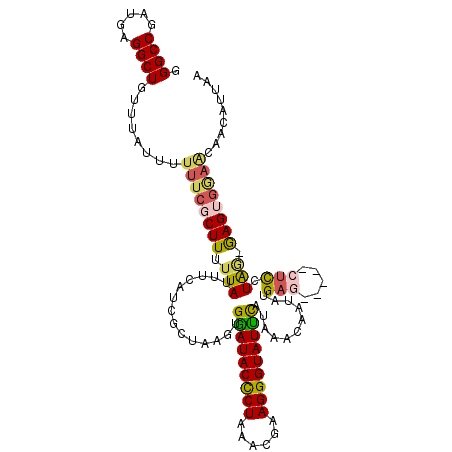
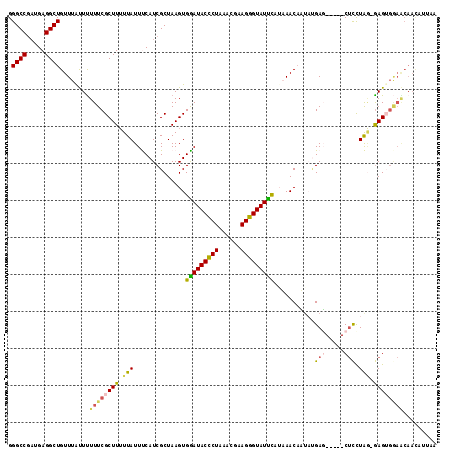
| Location | 1,405,904 – 1,406,010 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.25 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
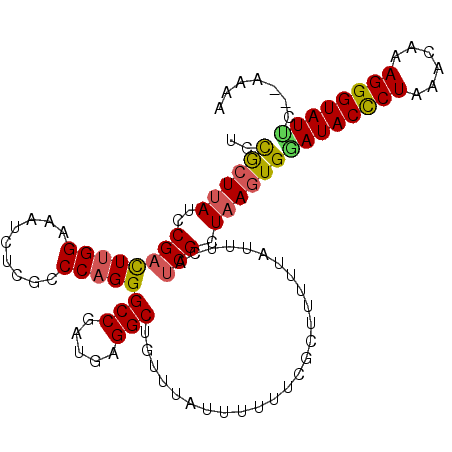
>2L_DroMel_CAF1 1405904 106 - 22407834 UUCGCUUAUCCGACUUGGAAAUCUCGCCCAGGGCCGAUGAGGCUGUUUAUUUUUUCGCUUUUUAUUUCAUCGCUAAGUGGAUACUCUAAGCGAAGGGUAUUU--UAAA (((((((((((.((((((.......(((...)))(((((((((.............))).......))))))))))))))))).....))))))........--.... ( -25.63) >DroSec_CAF1 24135 106 - 1 UCCGCUUAUCCGACUUGGAAAUCUCGCCCAGGGCCGAUGAGGCUGUUUAUUUUUUCGCUUUUUAUUUCAUCGCUAAGUGAAUACCCUAACCAAAGGGUAUUC--AUUA ....((((((.(.(((((.........))))).).))))))..................................((((((((((((......)))))))))--))). ( -28.10) >DroSim_CAF1 24323 106 - 1 UCCACUUAUCCGACUUGGAAAUCUCGCCCAGGGCCGAUGAGGCUGUUUAUUUUUUCGCUUUUUAUUUCAUCGCUAAGUGAAUACCCUAACCAAAGGGUAUUC--AUUA ....((((((.(.(((((.........))))).).))))))..................................((((((((((((......)))))))))--))). ( -28.40) >DroEre_CAF1 24095 103 - 1 UCCGCUUAUCCGACUUGGAAAUCUCGCCCAGGGCCGCUGAGGCUGUUUGCUUUUUCGCUU-UUAUUCCAUCGCUAAGUGGAUACCCUA-ACGCAGGGUAUCC---UAA ...(((((..(((..(((((.....((((((.....))).))).....((......))..-...)))))))).)))))(((((((((.-....)))))))))---... ( -33.50) >DroYak_CAF1 24469 106 - 1 GCCGCUUAUCCGACUUGGAAAUCUCGCCCAGGGCCAAUGAGGCUGUUUACUCUUUCGCUUUUUAUUCCAUCGCUAAGUGGAUACCCUAAACGCAGGGUAUCU--CAAA ..((((((..(((..(((((.....(((...)))....(((........)))............)))))))).))))))((((((((......)))))))).--.... ( -29.20) >DroAna_CAF1 24135 100 - 1 U-UGCUUAUCCGUUUCGGAAAUCUUGCCCAGGGCCGAUGAGGCUGUAUA-------GCUUUUGGUUUCAUCGCUUAAUGGAUACCCUAAACAAAGGGUAUAUUCGAUA .-............(((((........(((.(((.((((((((((....-------.....)))))))))))))...)))(((((((......))))))).))))).. ( -28.90) >consensus UCCGCUUAUCCGACUUGGAAAUCUCGCCCAGGGCCGAUGAGGCUGUUUAUUUUUUCGCUUUUUAUUUCAUCGCUAAGUGGAUACCCUAAACAAAGGGUAUUC__AAAA ..((((((..((((((((.........)))))(((.....)))..........................))).))))))((((((((......))))))))....... (-23.30 = -23.25 + -0.05)

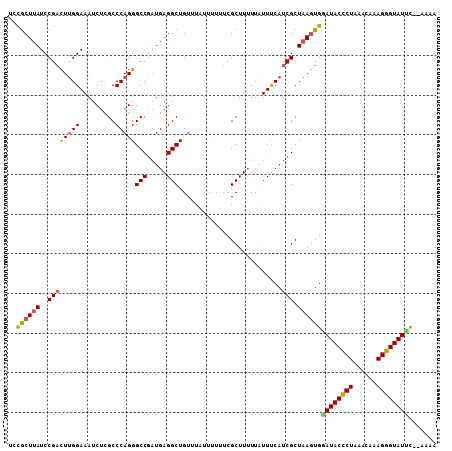
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:13 2006