
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,706,371 – 13,706,479 |
| Length | 108 |
| Max. P | 0.975589 |

| Location | 13,706,371 – 13,706,479 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -18.80 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
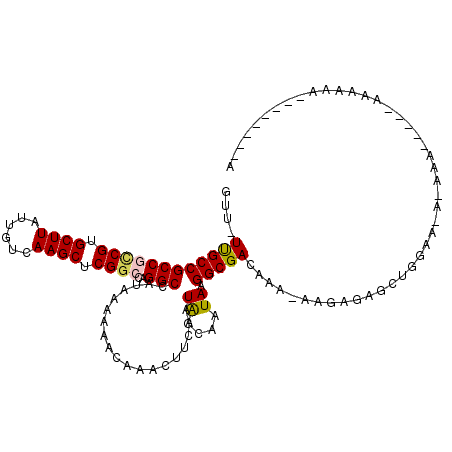
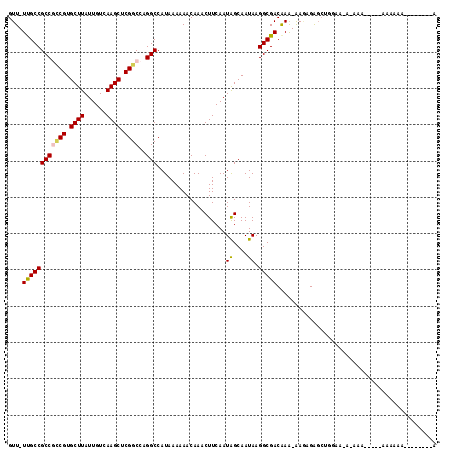
>2L_DroMel_CAF1 13706371 108 + 22407834 GUU-UUGCCGCCGCCGUGCUUAUUGUCAAGCUCGGCCAGGCCAUAAAAAACAAACUUCAAUGGCAAUAAGGCGACAAA-AAGAGAGCUUGAAUUAAAAAAAAAAAAAAA--------A .((-(((.((((((((.((((......)))).))))...(((((...............))))).....)))).))))-).............................--------. ( -28.26) >DroVir_CAF1 89714 107 + 1 GUU-UUGCCGCCAACGUGCUUAUUGUCAAGCUCGGCCAGGCCAUAAAAAACAAACUUCAAUAGCAAUAAGGCGACAAA-AAAUGGUA-GAAA---ACA-----ACAACAACGGUGCAA .((-((((((((..((.((((......)))).))....))).....................((......))......-....))))-))).---...-----............... ( -18.40) >DroPse_CAF1 74961 98 + 1 GUU-UCGCCGCCGCCGUGCUUAUUGUCAAGCUCGGCCAGGCCAUAAAAAACAAACUUCAAUAGCAAUAAGGCGACAAA-AA-AGAGC------A-AAAAAA--AAAAGA--------G ...-((((((((((((.((((......)))).))))..)))...................((....)).)))))....-..-.....------.-......--......--------. ( -23.60) >DroEre_CAF1 62904 113 + 1 GUU-UUGCCGCCGCCGUGCUUAUUGUCAAGCUCGGCCAGGCCAUAAAAAACAAACUUCAAUGGCAAUAAGGCGACAAA-AAGAGAGCUUGAAUACAAAAAACAAGGAAAAACA---AA .((-(((.((((((((.((((......)))).))))...(((((...............))))).....)))).))))-)......((((...........))))........---.. ( -29.36) >DroWil_CAF1 217451 92 + 1 GUUUUUGCCGCCAUCGUGCUUAUUGUCAAGCUCGGCCAGGCCAUAAAAAACAAACUUCAAUAGCAAUAAGGCGACGAAAAAAAGAGCAAGAA-------------------------- .((((((.((((...(.((((......)))).)(((...)))...........................)))).))))))............-------------------------- ( -18.50) >DroAna_CAF1 68817 102 + 1 GUU-UUGCCGCCGCCGUGCUUAUUGUCAAGCUCGGCCAGGCCAUAAAAAACAAACUUCAAUAGCAAUAAGGCGACAAG-GAGCGGGCUGGGAAAAAA------AUGCGA--------C .((-((.(((..(((.(((((.(((((......(((...)))....................((......))))))).-))))))))))).))))..------......--------. ( -26.40) >consensus GUU_UUGCCGCCGCCGUGCUUAUUGUCAAGCUCGGCCAGGCCAUAAAAAACAAACUUCAAUAGCAAUAAGGCGACAAA_AAGAGAGCUGGAA_A_AAA_____AAAAAA________A ....((((((((((((.((((......)))).))))..))).............((.....))......)))))............................................ (-18.80 = -18.83 + 0.03)

| Location | 13,706,371 – 13,706,479 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -27.39 |
| Energy contribution | -27.33 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
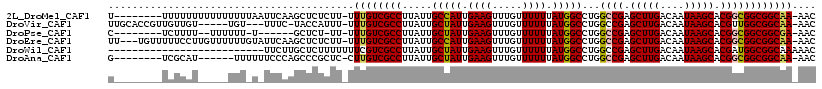
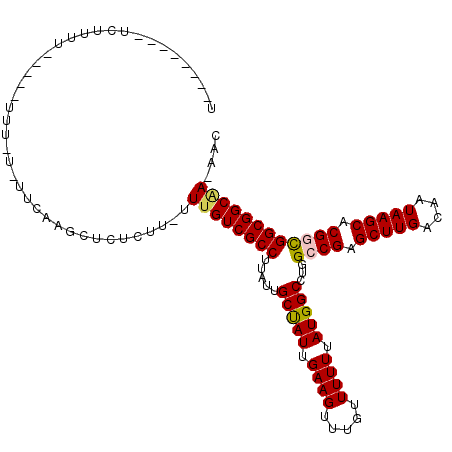
>2L_DroMel_CAF1 13706371 108 - 22407834 U--------UUUUUUUUUUUUUUUAAUUCAAGCUCUCUU-UUUGUCGCCUUAUUGCCAUUGAAGUUUGUUUUUUAUGGCCUGGCCGAGCUUGACAAUAAGCACGGCGGCGGCAA-AAC .--------.............................(-(((((((((.....(((((.((((.....)))).)))))...((((.(((((....))))).))))))))))))-)). ( -34.30) >DroVir_CAF1 89714 107 - 1 UUGCACCGUUGUUGU-----UGU---UUUC-UACCAUUU-UUUGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCCGAGCUUGACAAUAAGCACGUUGGCGGCAA-AAC ..(((.((....)).-----)))---....-.......(-(((((((((....((((((((((((((.........(((...)))))))))..)))).))))....))))))))-)). ( -25.00) >DroPse_CAF1 74961 98 - 1 C--------UCUUUU--UUUUUU-U------GCUCU-UU-UUUGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCCGAGCUUGACAAUAAGCACGGCGGCGGCGA-AAC .--------......--......-.------.....-.(-(((((((((.....(((((.((((.....)))).)))))...((((.(((((....))))).))))))))))))-)). ( -31.70) >DroEre_CAF1 62904 113 - 1 UU---UGUUUUUCCUUGUUUUUUGUAUUCAAGCUCUCUU-UUUGUCGCCUUAUUGCCAUUGAAGUUUGUUUUUUAUGGCCUGGCCGAGCUUGACAAUAAGCACGGCGGCGGCAA-AAC ..---........((((..(.....)..))))......(-(((((((((.....(((((.((((.....)))).)))))...((((.(((((....))))).))))))))))))-)). ( -35.30) >DroWil_CAF1 217451 92 - 1 --------------------------UUCUUGCUCUUUUUUUCGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCCGAGCUUGACAAUAAGCACGAUGGCGGCAAAAAC --------------------------...........(((((.((((((....((((((((((((((.........(((...)))))))))..)))).))))....))))))))))). ( -21.90) >DroAna_CAF1 68817 102 - 1 G--------UCGCAU------UUUUUUCCCAGCCCGCUC-CUUGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCCGAGCUUGACAAUAAGCACGGCGGCGGCAA-AAC .--------......------..................-.((((((((.....(((((.((((.....)))).)))))...((((.(((((....))))).))))))))))))-... ( -30.40) >consensus U________UCUUUU_____UUU_U_UUCAAGCUCUCUU_UUUGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCCGAGCUUGACAAUAAGCACGGCGGCGGCAA_AAC .........................................((((((((.....(((((.((((.....)))).)))))...((((.(((((....))))).)))))))))))).... (-27.39 = -27.33 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:40 2006