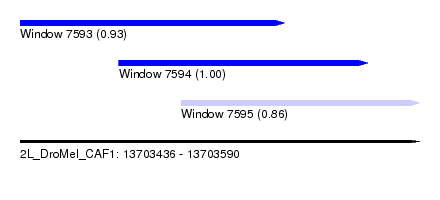
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,703,436 – 13,703,590 |
| Length | 154 |
| Max. P | 0.999737 |

| Location | 13,703,436 – 13,703,538 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -38.16 |
| Consensus MFE | -25.36 |
| Energy contribution | -26.24 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
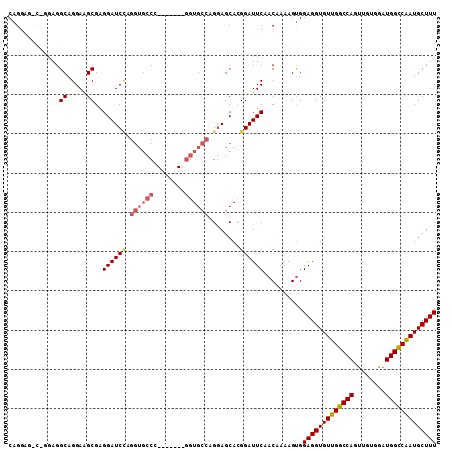
>2L_DroMel_CAF1 13703436 102 + 22407834 CAGGAGGCAGGAGGCAGGAGGCGAGGAUCCAGGUGCCC-------AGUGCCAGAAGCACGGAUUCAACAGAAGCGGAGGUGUUGGCCAGUUGUGGAUGGCCAAUGCUUU ......((.....((.....))..((((((.((...))-------.((((.....)))))))))).......)).(((((((((((((........))))))))))))) ( -34.80) >DroSec_CAF1 59737 109 + 1 CAGGAGACGGGAGGCAGGAAGCGAGGAUCCAGGUGCCCGGUGCCCGGUGCCGGGAGCACGGAUUCAACAAAAGUGGAGGUGUUGGCCAUUUGUGGAUGGCCAAUGCUUU ..(....).....((.....))..((((((..((.(((((..(...)..))))).))..))))))..........(((((((((((((((....))))))))))))))) ( -46.50) >DroSim_CAF1 58633 109 + 1 CAGGAGACGGGAGGCAGGAAGCGAGGAUCCAGGUGCCCGGUGCCCGGUGCCGGGAGCACGGAUUCUACAAAAGUGGAGGUGUUGGCCAUUUGUGGAUGGCCAAUGCUUU ..(....).....((.....)).(((((((..((.(((((..(...)..))))).))..))))))).........(((((((((((((((....))))))))))))))) ( -47.30) >DroEre_CAF1 60009 87 + 1 CAG--------AGGCAGGAGGCGAGGAUC-------CA-------GGUGGCAGGAGCAGGGAUUCGACGAAAGUGGAGGUGUUGGCCAGCAUUGGAUGGUCGAUGCUUU ..(--------(((((.....(((..(((-------((-------(.((((...((((....(((.((....)).))).)))).)))).)..)))))..))).)))))) ( -30.70) >DroYak_CAF1 60896 95 + 1 CAGG-------AGGCAGGAUGCGAGGAUCUAGGUGCCA-------GGUGCCAGGAGCAGGGAUUCCACGAGAGUGGAGGUGUUGGCCAGUCGUGGAUGGUCAAUGCUUU ....-------..((.....))..((((((..((.((.-------(....).)).))..))))))(((....)))(((((((((((((........))))))))))))) ( -31.50) >consensus CAGGAG_C_GGAGGCAGGAAGCGAGGAUCCAGGUGCCC_______GGUGCCAGGAGCACGGAUUCAACAAAAGUGGAGGUGUUGGCCAGUUGUGGAUGGCCAAUGCUUU .............((.....))..((((((.((((((........))))))........))))))..........(((((((((((((........))))))))))))) (-25.36 = -26.24 + 0.88)

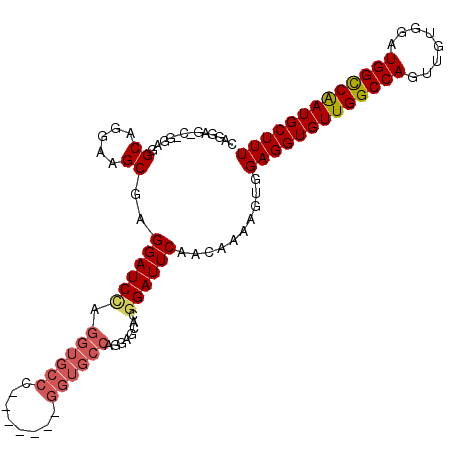
| Location | 13,703,474 – 13,703,570 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -31.16 |
| Energy contribution | -30.64 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
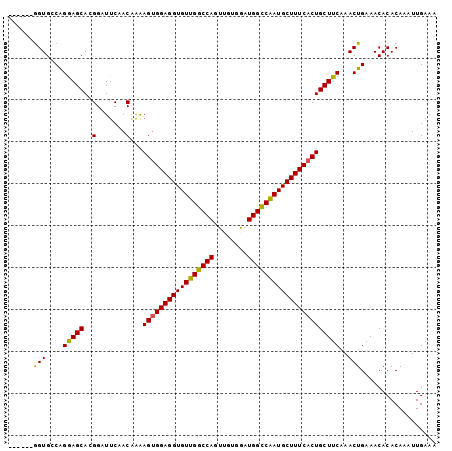
>2L_DroMel_CAF1 13703474 96 + 22407834 ------AGUGCCAGAAGCACGGAUUCAACAGAAGCGGAGGUGUUGGCCAGUUGUGGAUGGCCAAUGCUUUCACUGCUUCAAACUGAAACACACAAAUUGAAA ------.((((.....))))...((((...(((((((((((((((((((........))))))))))))...)))))))....))))............... ( -33.10) >DroSec_CAF1 59776 102 + 1 GUGCCCGGUGCCGGGAGCACGGAUUCAACAAAAGUGGAGGUGUUGGCCAUUUGUGGAUGGCCAAUGCUUUCACUGCUUCAAACUGAAACACACAAAUUGAAA (((((((....)))..))))...(((((....(((((((((((((((((((....)))))))))))))))))))(.(((.....))).).......))))). ( -40.80) >DroSim_CAF1 58672 102 + 1 GUGCCCGGUGCCGGGAGCACGGAUUCUACAAAAGUGGAGGUGUUGGCCAUUUGUGGAUGGCCAAUGCUUUCACUGCUUCAAACUGAAACACACAAAUUGAAA (((..((((....(((((..(.......)...(((((((((((((((((((....))))))))))))))))))))))))..))))...)))........... ( -40.30) >DroEre_CAF1 60032 96 + 1 ------GGUGGCAGGAGCAGGGAUUCGACGAAAGUGGAGGUGUUGGCCAGCAUUGGAUGGUCGAUGCUUUCACUGCUUCAAACUGAAACACACAAAUUGAAA ------.(((.(((((((((.......((....))((((((((((((((........)))))))))))))).))))))....)))...)))........... ( -34.60) >DroYak_CAF1 60927 96 + 1 ------GGUGCCAGGAGCAGGGAUUCCACGAGAGUGGAGGUGUUGGCCAGUCGUGGAUGGUCAAUGCUUUCACUGCUUCAAACUGAAACACACAAAUUGAAA ------.......((((((((((.(((((....)))))(((((((((((........)))))))))))))).)))))))....................... ( -35.00) >consensus ______GGUGCCAGGAGCACGGAUUCAACAAAAGUGGAGGUGUUGGCCAGUUGUGGAUGGCCAAUGCUUUCACUGCUUCAAACUGAAACACACAAAUUGAAA ......(((....(((((..(.......)...(((((((((((((((((........))))))))))))))))))))))..))).................. (-31.16 = -30.64 + -0.52)


| Location | 13,703,498 – 13,703,590 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -18.94 |
| Energy contribution | -20.48 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13703498 92 + 22407834 GAAGCGGAGGUGUUGGCCAGUUGUGGAUGGCCAAUGCUUUCACUGCUUCAAACUGAAACA--------CACAAAUUGAAAUAGAA--AACUUUUCCGAUGGA (((((((((((((((((((........))))))))))))...)))))))...........--------.....((((.((.((..--..)).)).))))... ( -27.40) >DroPse_CAF1 71755 93 + 1 AGCGUGGA-GUGGGGGCCAGCUGUGGAUGCC--------ACACUGCAUUGAGCUUAGUCAAGAAAUGCCAGAAAGUGAAAUAGAAGGAAGUUAGCGUGUGGA .((((...-((((.(.(((....))).).))--------)).(((((((...(((....))).)))).)))......................))))..... ( -19.20) >DroSec_CAF1 59806 92 + 1 AAAGUGGAGGUGUUGGCCAUUUGUGGAUGGCCAAUGCUUUCACUGCUUCAAACUGAAACA--------CACAAAUUGAAAUAGAA--AACUUUUCCGAUGGA ..(((((((((((((((((((....)))))))))))))))))))..(((((..((.....--------..))..)))))......--............... ( -29.90) >DroSim_CAF1 58702 92 + 1 AAAGUGGAGGUGUUGGCCAUUUGUGGAUGGCCAAUGCUUUCACUGCUUCAAACUGAAACA--------CACAAAUUGAAAUAGAA--AACUUUUCCGAUGGA ..(((((((((((((((((((....)))))))))))))))))))..(((((..((.....--------..))..)))))......--............... ( -29.90) >DroEre_CAF1 60056 92 + 1 AAAGUGGAGGUGUUGGCCAGCAUUGGAUGGUCGAUGCUUUCACUGCUUCAAACUGAAACA--------CACAAAUUGAAAUAGAA--AACUUUCCCGAUGGA ..(((((((((((((((((........)))))))))))))))))..(((((..((.....--------..))..)))))......--.....(((....))) ( -25.60) >DroYak_CAF1 60951 92 + 1 AGAGUGGAGGUGUUGGCCAGUCGUGGAUGGUCAAUGCUUUCACUGCUUCAAACUGAAACA--------CACAAAUUGAAAUAGAA--AACUUUCCCGAUGGA ..(((((((((((((((((........)))))))))))))))))..(((((..((.....--------..))..)))))......--.....(((....))) ( -26.00) >consensus AAAGUGGAGGUGUUGGCCAGUUGUGGAUGGCCAAUGCUUUCACUGCUUCAAACUGAAACA________CACAAAUUGAAAUAGAA__AACUUUUCCGAUGGA ..(((((((((((((((((........))))))))))))))))).......................................................... (-18.94 = -20.48 + 1.54)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:38 2006