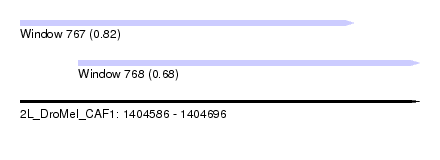
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,404,586 – 1,404,696 |
| Length | 110 |
| Max. P | 0.815081 |

| Location | 1,404,586 – 1,404,678 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -20.63 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
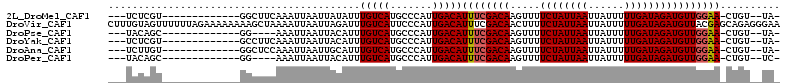
>2L_DroMel_CAF1 1404586 92 + 22407834 ---UCUCGU-------------GGCUUCAAAUUAAUUAUAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAA-CUGU--UA- ---....(.-------------(((..(((((.......)))))....))))..(((((((((((((.....((((((((......))))))))))))))))-.)))--))- ( -21.60) >DroVir_CAF1 42854 112 + 1 CUUUGUAGUUUUUUAGAAAAAAAAGCUAAAAUUAAUUAGAUUUGUCAUUCCCAUUGACAUUUCGACAACUUUUCUAUUAAUUAUUUUUGAUAGAUGUUACGAGCAGAGGGAA ((((((((((((((.....))))))))...........((..(((((.......)))))..))...(((...((((((((......)))))))).)))....)))))).... ( -24.60) >DroPse_CAF1 25316 88 + 1 ---UACAGC-------------GG----AAAUUAAUUACAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAA-CUGU--UA- ---...(((-------------((----..............(((((.......)))))((((((((.....((((((((......))))))))))))))))-))))--).- ( -20.20) >DroYak_CAF1 23099 92 + 1 ---UCUCGU-------------GCCUUCAAAUUAAUUACAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAA-CUGU--UA- ---....(.-------------((...(((((.......)))))....)).)..(((((((((((((.....((((((((......))))))))))))))))-.)))--))- ( -17.20) >DroAna_CAF1 22908 92 + 1 ---UCUUGU-------------GGCUCCAAAUUAAUUGCAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAA-CUGU--UA- ---...((.-------------(((..(((((.......)))))....))))).(((((((((((((.....((((((((......))))))))))))))))-.)))--))- ( -20.60) >DroPer_CAF1 25493 88 + 1 ---UACAGC-------------GG----AAAUUAAUUACAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAA-CUGU--UC- ---...(((-------------((----..............(((((.......)))))((((((((.....((((((((......))))))))))))))))-))))--).- ( -19.60) >consensus ___UCUAGU_____________GGCUUCAAAUUAAUUACAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAA_CUGU__UA_ ..........................................(((((.......)))))((((((((.....((((((((......)))))))))))))))).......... (-14.90 = -15.10 + 0.20)
| Location | 1,404,602 – 1,404,696 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -17.68 |
| Consensus MFE | -14.77 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
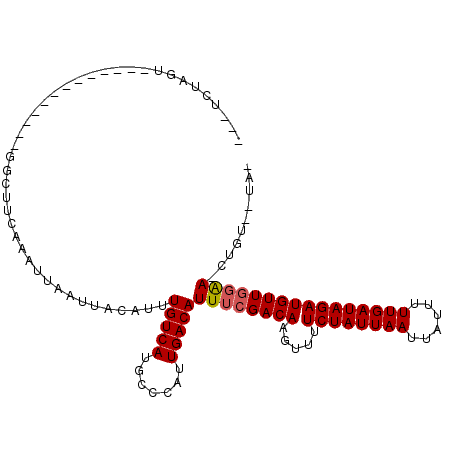
>2L_DroMel_CAF1 1404602 94 + 22407834 UAAUUAUAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAACUGUUAAAAAUACAACCCG-----------------AAAGC .....................((((((((((((((.....((((((((......)))))))))))))))).))))))..........(.-----------------...). ( -17.20) >DroPse_CAF1 25328 104 + 1 UAAUUACAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAACUGUUAAAAAUACAACCGA-------AAAAAACCGAAAAGC ..........(((((.......)))))(((((.....(((((.......((((((((((((.........)))))))))))).....))-------)))....)))))... ( -17.20) >DroEre_CAF1 22835 93 + 1 UAAUUACAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAACUGUUAAAAAUACAACCCG-----------------A-AGC .....................((((((((((((((.....((((((((......)))))))))))))))).))))))............-----------------.-... ( -16.90) >DroWil_CAF1 25367 79 + 1 UAAUUAGAUUUGUCAUUUCCAUUGACAUUUCGACAACUUUUCUAUUAAUUAUUUUUGAUAGAUGUGGUAA-------GAAA-------------------------AAAGC ......((..(((((.......)))))..))....(((..((((((((......))))))))...)))..-------....-------------------------..... ( -16.00) >DroAna_CAF1 22924 111 + 1 UAAUUGCAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAACUGUUAAAAAUACAAUCCGGACAACAAAAAUGCUUUAAAGC .....(((((((((.......((((((((((((((.....((((((((......)))))))))))))))).)))))).....(.....))))).....)))))........ ( -22.10) >DroPer_CAF1 25505 104 + 1 UAAUUACAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAACUGUUCAAAAUACAACCGA-------AAAAAACCGAAAAGC ..........(((((.......)))))(((((........((((((((......))))))))..((((...(((.......))).))))-------.......)))))... ( -16.70) >consensus UAAUUACAUUUGUCAUGCCCAUUGACAUUUCGACAAGUUUUCUAUUAAUUAUUUUUGAUAGAUGUUGGAACUGUUAAAAAUACAACCCG_________________AAAGC ..........(((((.......)))))((((((((.....((((((((......))))))))))))))))......................................... (-14.77 = -15.10 + 0.33)

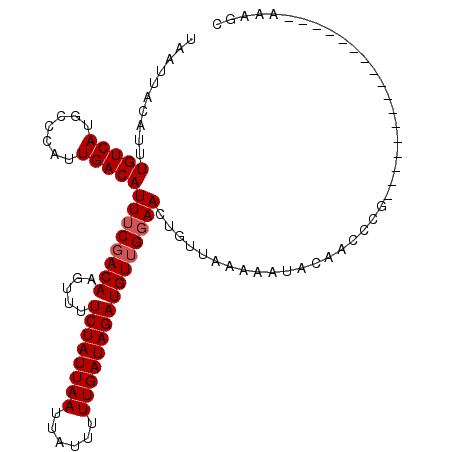
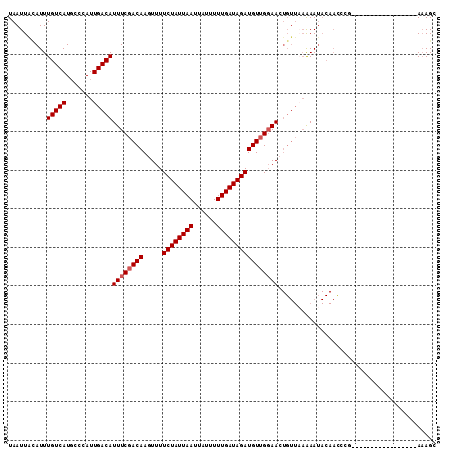
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:09 2006