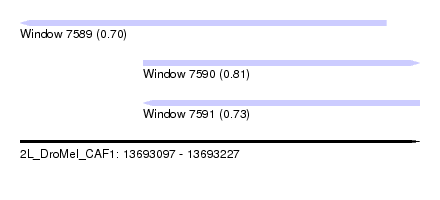
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,693,097 – 13,693,227 |
| Length | 130 |
| Max. P | 0.808126 |

| Location | 13,693,097 – 13,693,216 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.34 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13693097 119 - 22407834 -CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGACCAUAAAAAGCGAUAACAUCAUAUUUAUA -....(((.(................((((.....))))(((.(((((((((.............).)))))))).))).......).)))............................. ( -18.42) >DroVir_CAF1 70132 120 - 1 ACCAAUCCCCGAGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACAGAAGACUGAUUGCUGGCAAUAAAAAUGGACCAUAAAAAGCGAUAACAUCAUAUUUAUA .(((......................((((.....))))((.((((((((((.............).))))))))).))........))).............................. ( -18.62) >DroGri_CAF1 74460 120 - 1 ACCAAUCCCCUAGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUGAGCAAACAAAACAGAAGACUGAUUGCUGGCAAUAAAAAUGGACCAUAAAAAGCGAUAACAUCAUAUUUAUA .(((......................((((.....))))((.((((((.(((.............).)).)))))).))........))).............................. ( -14.52) >DroMoj_CAF1 81338 120 - 1 GCCGAUCUCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCUGGCAAUCAGCAAACAAAACAGGAGACUGAUUGCUGGCAAUAAAAAUGGACCAUAAAAAGCGAUAACAUCAUAUUUAUA .(((.....)))..............(((((........((..((((((((....(......)....))))))))..)).....)))))............................... ( -23.72) >DroAna_CAF1 53943 119 - 1 -CCAAUCCCCGGGAAUUAAACAAACACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCAAUAAAAGUGGACCAUAAAAAGCGAUAACAUCAUAUUUAUA -....(((.(................((((.....))))((..(((((((((.............).))))))))..)).......).)))............................. ( -17.22) >DroPer_CAF1 60386 118 - 1 -CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCUAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCAAUAAAAAUGGACCAUAA-AAGCGAUAACAUCAUAUUUAUA -(((.(((...)))............((((.....))))((..(((((((((.............).))))))))..))........))).......-...................... ( -18.82) >consensus _CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACACAAGACUGAUUGCCGGCAAUAAAAAUGGACCAUAAAAAGCGAUAACAUCAUAUUUAUA .(((......................((((.....))))((..(((((((((.............).))))))))..))........))).............................. (-16.31 = -16.34 + 0.03)


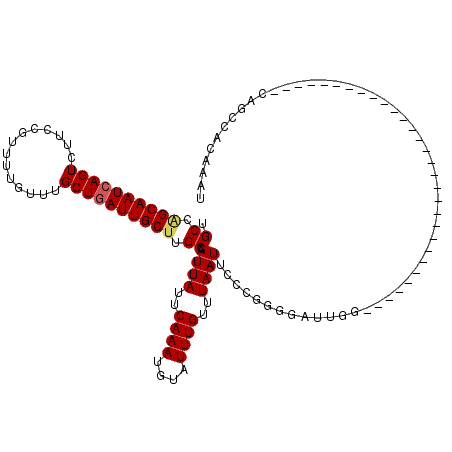
| Location | 13,693,137 – 13,693,227 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.69 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13693137 90 + 22407834 UGCAGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG------------------------------CAGCCACAAAU .(((((((((((((...((.....))..)))))))))))))...........(((..(((((.(((((....)))))))------------------------------)))..)))... ( -28.00) >DroVir_CAF1 70172 120 + 1 UGCCAGCAAUCAGUCUUCUGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCUCGGGGAUUGGUUCCGCUGCCAAUGCCGCUGCUCCGCUGGGCAGGCACAAAU ((((((((((((((...(......)...))))))))))((((((((..((((....))))..)))).....((.((((((......)))))).))((......))..))))))))..... ( -39.30) >DroGri_CAF1 74500 113 + 1 UGCCAGCAAUCAGUCUUCUGUUUUGUUUGCUCAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCUAGGGGAUUGGU--CACUG-----ACCGCUGGUCCGCUGGGCGUUCACAAAU ..(((((.((((((((((((....((..((.....))..)).((((..((((....))))..))))..))))))))))))--....(-----(((...)))).)))))............ ( -29.50) >DroMoj_CAF1 81378 102 + 1 UGCCAGCAAUCAGUCUCCUGUUUUGUUUGCUGAUUGCCAGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGAGAUCGGC------------------GCUGGGCUGGGCGAGCACAAAU .(((.(((((((((...(......)...)))))))))(((((((((..((((....))))..))))..(((.....))).------------------....)))))))).......... ( -30.40) >DroAna_CAF1 53983 90 + 1 UGCCGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUGUUUGUUUAAUUCCCGGGGAUUGG------------------------------CGGCCACAAAU .((.((((((((((...((.....))..)))))))))).))...........((((..((((.(((((....)))))))------------------------------))..))))... ( -27.00) >DroPer_CAF1 60425 90 + 1 UGCCGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUAGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG------------------------------CAGCCACAAAU .((..(((((((((...((.....))..)))))))))..))...........(((..(((((.(((((....)))))))------------------------------)))..)))... ( -24.70) >consensus UGCCAGCAAUCAGUCUUCCGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG______________________________CAGCCACAAAU .((.((((((((((..............)))))))))).)).((((..((((....))))..))))...................................................... (-15.52 = -15.69 + 0.17)


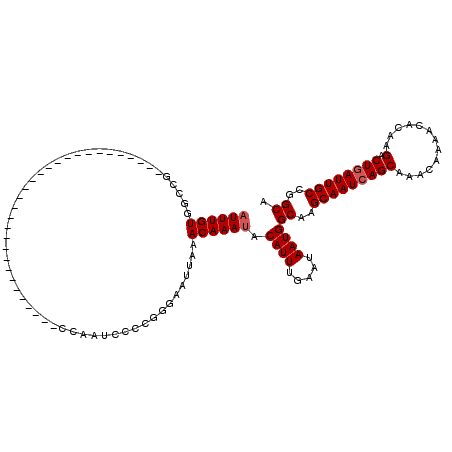
| Location | 13,693,137 – 13,693,227 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -13.65 |
| Energy contribution | -13.99 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13693137 90 - 22407834 AUUUGUGGCUG------------------------------CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCA ((((((.....------------------------------((........))......)))))).((((.....))))(((.(((((((((.............).)))))))).))). ( -20.52) >DroVir_CAF1 70172 120 - 1 AUUUGUGCCUGCCCAGCGGAGCAGCGGCAUUGGCAGCGGAACCAAUCCCCGAGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACAGAAGACUGAUUGCUGGCA (((((((((((((..((......))))))..)))..(((.........)))........)))))).((((.....))))((.((((((((((.............).))))))))).)). ( -32.32) >DroGri_CAF1 74500 113 - 1 AUUUGUGAACGCCCAGCGGACCAGCGGU-----CAGUG--ACCAAUCCCCUAGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUGAGCAAACAAAACAGAAGACUGAUUGCUGGCA .........(((...)))(.((((((((-----(((((--.(((.((.....))......((((....)))).....))))..((.....))..............))))))))))))). ( -24.70) >DroMoj_CAF1 81378 102 - 1 AUUUGUGCUCGCCCAGCCCAGC------------------GCCGAUCUCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCUGGCAAUCAGCAAACAAAACAGGAGACUGAUUGCUGGCA ......(((.....)))(((((------------------((.(((..((((..((((..((((....))))..))))..))))..))).))........(((....)))...))))).. ( -28.00) >DroAna_CAF1 53983 90 - 1 AUUUGUGGCCG------------------------------CCAAUCCCCGGGAAUUAAACAAACACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCA .......((((------------------------------(((.(((...)))......((((....)))).....))))..(((((((((.............).)))))))).))). ( -22.62) >DroPer_CAF1 60425 90 - 1 AUUUGUGGCUG------------------------------CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCUAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCA ((((((.....------------------------------((........))......)))))).((((.....))))((..(((((((((.............).))))))))..)). ( -21.02) >consensus AUUUGUGGCCG______________________________CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACACAAGACUGAUUGCCGGCA ((((((.....................................................)))))).((((.....))))((..(((((((((.............).))))))))..)). (-13.65 = -13.99 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:34 2006