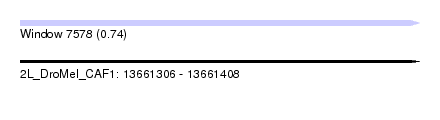
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,661,306 – 13,661,408 |
| Length | 102 |
| Max. P | 0.741893 |

| Location | 13,661,306 – 13,661,408 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.50 |
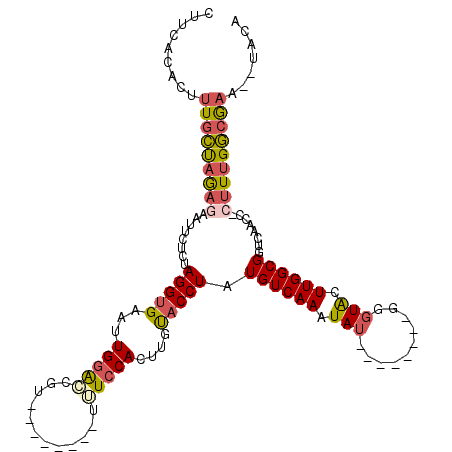
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -16.77 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
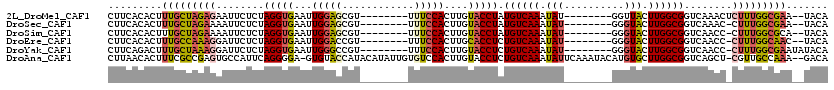
>2L_DroMel_CAF1 13661306 102 + 22407834 CUUCACACUUUGCUAGAGAAUUCUCUAGGUGAAUUGGAGCGU--------UUUCCACUUGUACCUAUGUCAAAUAU--------GGUUACUUGGCGGUCAAACUCUUUGGCGAA--UACA ........(((((((((((......((((((...(((((...--------.)))))....))))))((((((.((.--------...)).)))))).......)))))))))))--.... ( -26.70) >DroSec_CAF1 17777 101 + 1 CUUCACACUUUGCUAGAAAAUUCUCUAGGUGAAUUGGAGCGU--------UUUCCACUUGUACCUAUGUCAAAUAU--------GGGUACUUGGCGGUCAAAC-CUUUGGCGAA--UACA .(((......(((((((((((.((((((.....)))))).))--------)))).....((((((((((...))))--------)))))).)))))(((((..-..))))))))--.... ( -29.10) >DroSim_CAF1 17531 101 + 1 CUUCACACUUUGCUAGAAAAUUCUCUAGGUGAAUUGGAGCGU--------UUUCCACUUGUACCUAUGUCAAAUAU--------GGGUACUUGGCGGUCAACC-CUUUGGCGCA--UACA ............(((((......)))))(((..(..(((.((--------(..((.((.((((((((((...))))--------))))))..)).))..))).-)))..)..))--)... ( -29.10) >DroEre_CAF1 17133 101 + 1 CUUCACACUUUGCCAAAGGAUUCUCUAGGUGAAUUGGACCGU--------UUUCCACUUGCACCUCUGUCAAAUAU--------GGGUACUUGGCGGUCAACC-CUUUGGCAAC--UACA .........((((((((((.......(((((((.((((....--------..)))).)).)))))(((((((.((.--------...)).))))))).....)-))))))))).--.... ( -31.70) >DroYak_CAF1 17245 103 + 1 CUUCAGACUUUGCUAAAGGAUUCUCUAGGUGAAUUGGGCCGU--------UUUCCACUUGUACCUCUGUCAAAUAU--------GGGUACUUGGCGGUCAACC-CUUUGGCGAAUAUACA ........(((((((((((((((.......))))..(((((.--------...(((...((((((.(((...))).--------)))))).))))))))...)-))))))))))...... ( -30.90) >DroAna_CAF1 19833 116 + 1 CUUAACACUUUCGCCGAGUGCCAUUCAGGGGA-GUGUACCAUACAUAUUGUGUCCACUUGUACCUCUGUCAAAUAUUCAAAUACAUGUGCUUGGCGGUCAGCU-CGUUGCCAAA--GACA ...........(((((((..(....(((((((-(((.((((.......)).)).))))....))))))..................)..)))))))(((.((.-....))....--))). ( -28.65) >consensus CUUCACACUUUGCUAGAGAAUUCUCUAGGUGAAUUGGACCGU________UUUCCACUUGUACCUAUGUCAAAUAU________GGGUACUUGGCGGUCAACC_CUUUGGCGAA__UACA .........(((((((((........(((((...(((((............)))))....))))).((((((.(((..........))).))))))........)))))))))....... (-16.77 = -17.08 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:22 2006