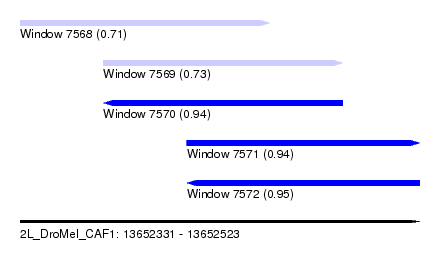
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,652,331 – 13,652,523 |
| Length | 192 |
| Max. P | 0.954340 |

| Location | 13,652,331 – 13,652,451 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.59 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -16.03 |
| Energy contribution | -17.15 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13652331 120 + 22407834 CUUGGCGGUCAAACUCUUUGGCGAAUACAUCUCCAUCAUUCGCAGAUACAUUUGAACGUUUCAAUUUUCAAGUGUUGAGAUUGGCCCACUUGGAAGUGGCCUGAUGUUUCCUUCAAGACA ...(((((((((.(((....((((((...........))))))....(((((((((.((....)).))))))))).))).))))))((((....)))))))...(((((......))))) ( -32.30) >DroPse_CAF1 20769 106 + 1 CUUGGCGGUCAG-------------GACAUCUACAUCAAUCGCACUCCCAUUUAAAGGUUUCAAUUUUCGAGUGCGCCUAUUGGCCCACUUCUGGG-CUCUUGAUGUCUCUUCCAAGACA (((((.((...(-------------((((((.....(((((((((((((.......))...........)))))))...))))(((((....))))-)....))))))))).)))))... ( -33.90) >DroPer_CAF1 21094 106 + 1 CUUGGCGGUCAG-------------GACAUCUACAUCAGUCGCACUCCCAUUUAAAGGUUUCAAUUUUCGAGUGCGCCUAUUGGCCCACUUCUGGG-CUCUUGAUGUCUCUUCCAAGACA (((((.((...(-------------((((((..((..((.(((((((((.......))...........))))))).))..))(((((....))))-)....))))))))).)))))... ( -36.00) >consensus CUUGGCGGUCAG_____________GACAUCUACAUCAAUCGCACUCCCAUUUAAAGGUUUCAAUUUUCGAGUGCGCCUAUUGGCCCACUUCUGGG_CUCUUGAUGUCUCUUCCAAGACA (((((.((.................((((((....((((((((((((......................)))))))...)))))(((......)))......)))))).)).)))))... (-16.03 = -17.15 + 1.12)

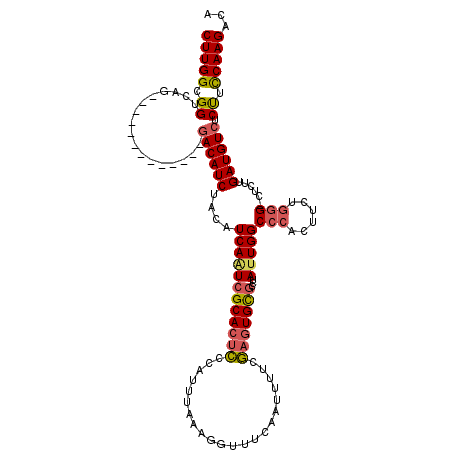
| Location | 13,652,371 – 13,652,486 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -21.05 |
| Energy contribution | -19.28 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13652371 115 + 22407834 CGCAGAUACAUUUGAACGUUUCAAUUUUCAAGUGUUGAGAUUGGCCCACUUGGAAGUGGCCUGAUGUUUCCUUCAAGACAGCUUUCAGCUUUGUGCCGGCCACAUUCCACU-----CGGC .......(((((((((.((....)).)))))))))........(((....(((((((((((...(((((......)))))((.....))........)))))).)))))..-----.))) ( -30.80) >DroPse_CAF1 20796 119 + 1 CGCACUCCCAUUUAAAGGUUUCAAUUUUCGAGUGCGCCUAUUGGCCCACUUCUGGG-CUCUUGAUGUCUCUUCCAAGACAACUCAAAGCCUUGUGUUGGCCACAUUCGUCUUGGCUUGGC .((((((((.......))...........))))))(((...(((((.((....(((-((.(((((((((......)))))..)))))))))...)).))))).....((....))..))) ( -36.40) >DroPer_CAF1 21121 119 + 1 CGCACUCCCAUUUAAAGGUUUCAAUUUUCGAGUGCGCCUAUUGGCCCACUUCUGGG-CUCUUGAUGUCUCUUCCAAGACAACUCAAAGCCUUGUGUUGGCCACAUUCGUCUUGGCUUGGC .((((((((.......))...........))))))(((...(((((.((....(((-((.(((((((((......)))))..)))))))))...)).))))).....((....))..))) ( -36.40) >consensus CGCACUCCCAUUUAAAGGUUUCAAUUUUCGAGUGCGCCUAUUGGCCCACUUCUGGG_CUCUUGAUGUCUCUUCCAAGACAACUCAAAGCCUUGUGUUGGCCACAUUCGUCUUGGCUUGGC .((((((......................))))))((..((((((((((....(((......(.(((((......))))).)......))).)))..))))).))..))........... (-21.05 = -19.28 + -1.77)


| Location | 13,652,371 – 13,652,486 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -21.72 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.937978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13652371 115 - 22407834 GCCG-----AGUGGAAUGUGGCCGGCACAAAGCUGAAAGCUGUCUUGAAGGAAACAUCAGGCCACUUCCAAGUGGGCCAAUCUCAACACUUGAAAAUUGAAACGUUCAAAUGUAUCUGCG (((.-----(.(((((.((((((((((....((.....)))))).(((.(....).))))))))))))))..).)))........(((.(((((..........))))).)))....... ( -34.70) >DroPse_CAF1 20796 119 - 1 GCCAAGCCAAGACGAAUGUGGCCAACACAAGGCUUUGAGUUGUCUUGGAAGAGACAUCAAGAG-CCCAGAAGUGGGCCAAUAGGCGCACUCGAAAAUUGAAACCUUUAAAUGGGAGUGCG (((...(((((((((....((((.......)))).....)))))))))............(.(-((((....))))))....)))((((((...........((.......)))))))). ( -41.30) >DroPer_CAF1 21121 119 - 1 GCCAAGCCAAGACGAAUGUGGCCAACACAAGGCUUUGAGUUGUCUUGGAAGAGACAUCAAGAG-CCCAGAAGUGGGCCAAUAGGCGCACUCGAAAAUUGAAACCUUUAAAUGGGAGUGCG (((...(((((((((....((((.......)))).....)))))))))............(.(-((((....))))))....)))((((((...........((.......)))))))). ( -41.30) >consensus GCCAAGCCAAGACGAAUGUGGCCAACACAAGGCUUUGAGUUGUCUUGGAAGAGACAUCAAGAG_CCCAGAAGUGGGCCAAUAGGCGCACUCGAAAAUUGAAACCUUUAAAUGGGAGUGCG ..................(((((.((.((((((........))))))...((....)).............)).))))).....(((((((...(.((((.....)))).)..))))))) (-21.72 = -22.50 + 0.78)

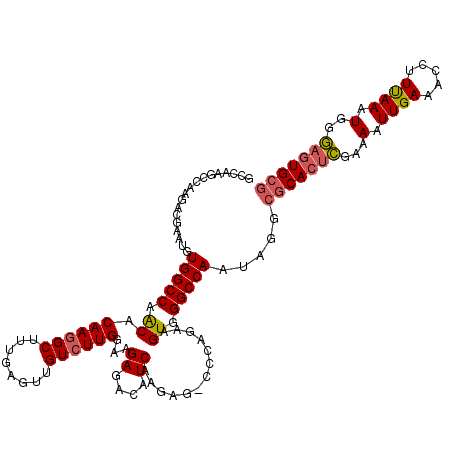
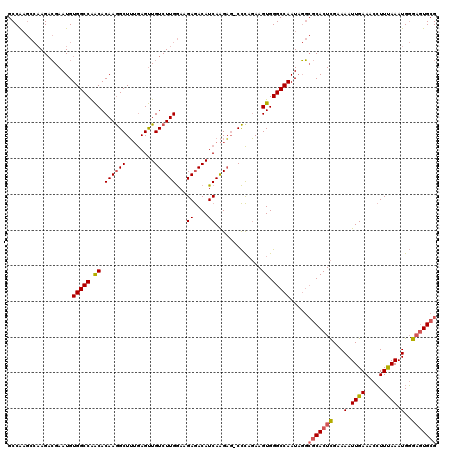
| Location | 13,652,411 – 13,652,523 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -42.33 |
| Consensus MFE | -25.12 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13652411 112 + 22407834 UUGGCCCACUUGGAAGUGGCCUGAUGUUUCCUUCAAGACAGCUUUCAGCUUUGUGCCGGCCACAUUCCACU-----CGGCCAGAU-UGCUC--CGUGUGAAGGGCCAUAUAGCUGCAAUC .(((((....(((((((((((...(((((......)))))((.....))........)))))).)))))..-----.)))))(((-(((.(--..((((......))))..)..)))))) ( -39.60) >DroPse_CAF1 20836 119 + 1 UUGGCCCACUUCUGGG-CUCUUGAUGUCUCUUCCAAGACAACUCAAAGCCUUGUGUUGGCCACAUUCGUCUUGGCUUGGCCAGUUGUGCUCACAGUGUGAAGUGCCACACUGGUGCAACC .((((((((....(((-((.(((((((((......)))))..))))))))).)))..(((((.........))))).)))))((((..(...(((((((......))))))))..)))). ( -45.90) >DroPer_CAF1 21161 119 + 1 UUGGCCCACUUCUGGG-CUCUUGAUGUCUCUUCCAAGACAACUCAAAGCCUUGUGUUGGCCACAUUCGUCUUGGCUUGGCCAGUUGUGCUCACAGUGUGAAGUGCCACACUGGUGGAACC .((((((((....(((-((.(((((((((......)))))..))))))))).)))..(((((.........))))).)))))(((.(((...(((((((......)))))))))).))). ( -41.50) >consensus UUGGCCCACUUCUGGG_CUCUUGAUGUCUCUUCCAAGACAACUCAAAGCCUUGUGUUGGCCACAUUCGUCUUGGCUUGGCCAGUUGUGCUCACAGUGUGAAGUGCCACACUGGUGCAACC ((((((.......(((......(.(((((......))))).)......)))((((.....)))).............))))))...(((...(((((((......)))))))..)))... (-25.12 = -24.80 + -0.32)

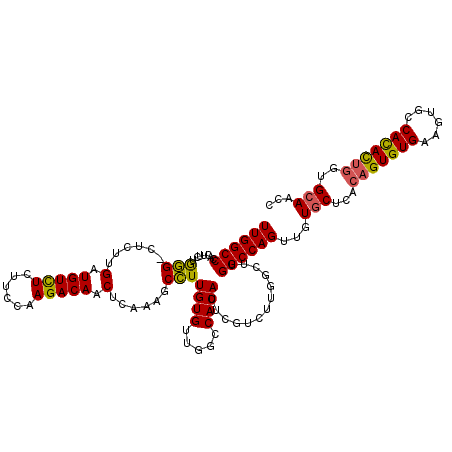
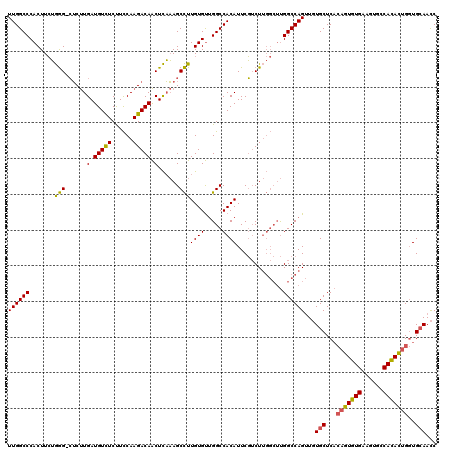
| Location | 13,652,411 – 13,652,523 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -44.47 |
| Consensus MFE | -23.98 |
| Energy contribution | -25.77 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13652411 112 - 22407834 GAUUGCAGCUAUAUGGCCCUUCACACG--GAGCA-AUCUGGCCG-----AGUGGAAUGUGGCCGGCACAAAGCUGAAAGCUGUCUUGAAGGAAACAUCAGGCCACUUCCAAGUGGGCCAA ((((((.(((....)))((.......)--).)))-)))(((((.-----(.(((((.((((((((((....((.....)))))).(((.(....).))))))))))))))..).))))). ( -46.50) >DroPse_CAF1 20836 119 - 1 GGUUGCACCAGUGUGGCACUUCACACUGUGAGCACAACUGGCCAAGCCAAGACGAAUGUGGCCAACACAAGGCUUUGAGUUGUCUUGGAAGAGACAUCAAGAG-CCCAGAAGUGGGCCAA ((((...((((((((.(((........)))..))).)))))...))))..........(((((.((....((((((....((((((....))))))...))))-)).....)).))))). ( -43.20) >DroPer_CAF1 21161 119 - 1 GGUUCCACCAGUGUGGCACUUCACACUGUGAGCACAACUGGCCAAGCCAAGACGAAUGUGGCCAACACAAGGCUUUGAGUUGUCUUGGAAGAGACAUCAAGAG-CCCAGAAGUGGGCCAA (((.((((((((((((....)))))))).........((((.(...(((((((((....((((.......)))).....)))))))))..((....))....)-.))))..))))))).. ( -43.70) >consensus GGUUGCACCAGUGUGGCACUUCACACUGUGAGCACAACUGGCCAAGCCAAGACGAAUGUGGCCAACACAAGGCUUUGAGUUGUCUUGGAAGAGACAUCAAGAG_CCCAGAAGUGGGCCAA ...(((.(((((((((....)))))))).).)))....(((((.............((((.....))))..(((((......((((((........))))))......))))).))))). (-23.98 = -25.77 + 1.78)

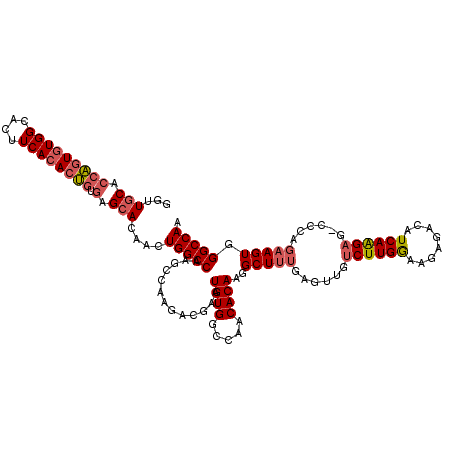
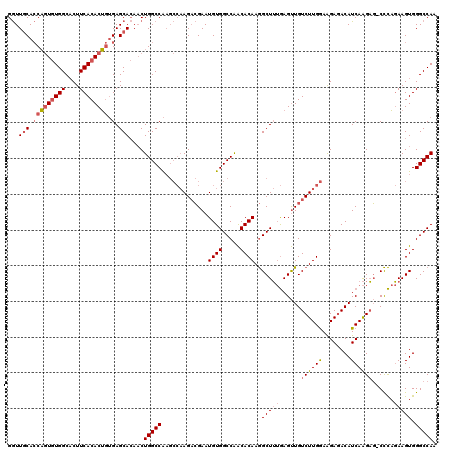
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:16 2006