
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,651,161 – 13,651,462 |
| Length | 301 |
| Max. P | 0.995362 |

| Location | 13,651,161 – 13,651,263 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.19 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -15.34 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13651161 102 + 22407834 ------UUUUGACAUUCUAUCCGAGUAUUCAUUGUGCAGCAGCAUACAACCACAUUUGCUGU------------GGUAGCUCUGGUUUAUGAAUGUUGAUAAAUAUUUACAAUUUUAAUA ------..(..((((((...((((((......(((((....)))))..((((((.....)))------------))).)))).)).....))))))..)..................... ( -25.90) >DroPse_CAF1 19738 119 + 1 UCGAGCCUUUUAUAUUUAUUACGAAAUUCCAUUAUGCGCAUUCAUGCAUCUAAAUUUUUUUCCUUCAGGAGGGGAGGGGCUUUGGCUCAUGAAUGUUGAUAAAUAUUUCCAAU-UUAAUA ...........((((((((((.(......).......(((((((((...(((((.((((((((((....)))))))))).)))))..))))))))))))))))))).......-...... ( -25.10) >DroSim_CAF1 16392 102 + 1 ------CUUUGACAUUCUAUCCGAGUAUUCAUUGUGCAGCACCACACAACCACAUUUGCUGC------------GGUAGCUCUGGUUUAUGAAUGUUGAUAAAUAUUUACAAUUUUAAUA ------..(..((((((...(((.((((.....))))((((((.(((((......))).)).------------))).))).))).....))))))..)..................... ( -20.40) >DroPer_CAF1 20063 119 + 1 UCAAGCCUUUUAUAUUUAUUACGAAAUUCCAUUAUGCGCAUUCAUGCAUCUAAAUUUUUUCCCUUCAGGAAGGGAGGGGCUUUGGCUCAUGAAUGUUGAUAAAUAUUUCCAAU-UUAAUA ...........((((((((((.(......).......(((((((((...(((((.((((((((((....)))))))))).)))))..))))))))))))))))))).......-...... ( -28.30) >consensus ______CUUUGACAUUCAAUACGAAAAUCCAUUAUGCACAAUCAUACAACCAAAUUUGCUGC____________AGGAGCUCUGGCUCAUGAAUGUUGAUAAAUAUUUACAAU_UUAAUA ........(((((((((...............((((......)))).(((((((.((((((((((....)))))))))).)))))))...)))))))))..................... (-15.34 = -15.40 + 0.06)

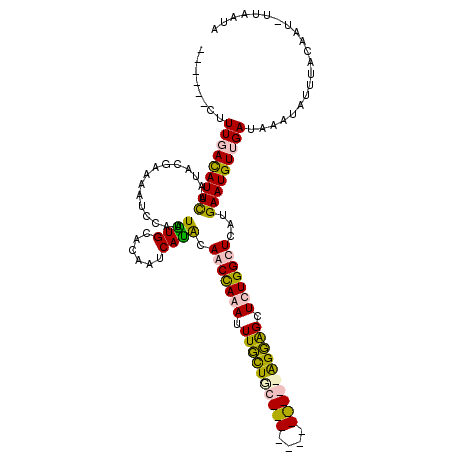
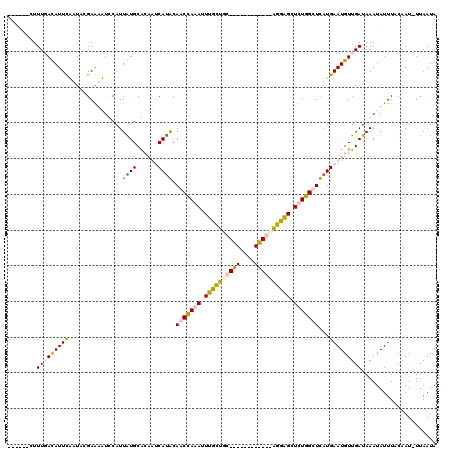
| Location | 13,651,263 – 13,651,382 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -16.10 |
| Energy contribution | -16.85 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13651263 119 - 22407834 AACAAAACUUUUGCGUUAA-AAACAGAAAAGUGUUUUGAGCUACUCGACACUAGGCAAAAAACGCCAGGAAAAGUGCAAAAAAAAAGUAAACACAGUCUGAAAGUAGCCAACUAAACUUU ......((((((..((...-..))..))))))...(((.((((((.(((.((.(((.......))).))....(((...............))).)))....)))))))))......... ( -19.66) >DroPse_CAF1 19857 99 - 1 AACAAAACUUUCGCGUUAAAAAACAGAAAAGUGUUUUGUGCUCCUCGACACAA------------CAAGAGGAGUGCAGAAAAA---------UGGUCGGAGAGUGGCCAACUAAACUUU ........((((..(((....))).))))(((.(((((..((((((.......------------...))))))..)))))...---------((((((.....)))))))))....... ( -27.10) >DroSim_CAF1 16494 119 - 1 AACAAAACUUUUGCGUUAA-AAACAGAAAAGUGUUUUGAGCUACUCGACACUAGGCAAAAAACGCCAGGAAAAGUGCAAAAAAAAAGUAAACACAGUCUGAAAGUAGCCAACUAAACUUU ......((((((..((...-..))..))))))...(((.((((((.(((.((.(((.......))).))....(((...............))).)))....)))))))))......... ( -19.66) >DroPer_CAF1 20182 99 - 1 AACAAAACUUUCGCGUUAAAAAACAGAAAAGUGUUUUGUGCUCCUCGACACAA------------CAAGAGGAGUGCAGAAAAA---------UGGUCGGAGAGUGGCCAACUAAACUUU ........((((..(((....))).))))(((.(((((..((((((.......------------...))))))..)))))...---------((((((.....)))))))))....... ( -27.10) >consensus AACAAAACUUUCGCGUUAA_AAACAGAAAAGUGUUUUGAGCUACUCGACACAA____________CAAGAAAAGUGCAAAAAAA_________CAGUCGGAAAGUAGCCAACUAAACUUU ......((((((..(((....))).........(((((((((((((......................)))))))))))))..................))))))............... (-16.10 = -16.85 + 0.75)

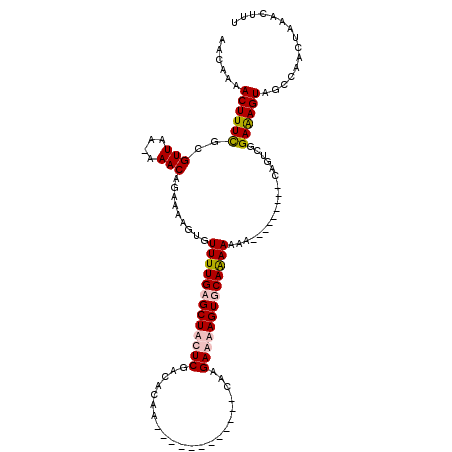
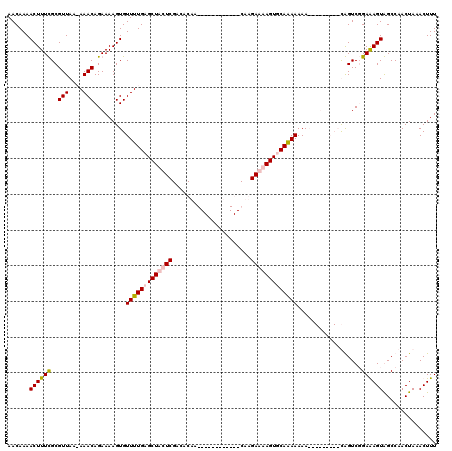
| Location | 13,651,303 – 13,651,422 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.29 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -18.90 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13651303 119 + 22407834 UUUGCACUUUUCCUGGCGUUUUUUGCCUAGUGUCGAGUAGCUCAAAACACUUUUCUGUUU-UUAACGCAAAAGUUUUGUUAUGGAAAUGCAGCAAGUUCUUGGUACUUUUUCAUAUUUUU ...((((((..(((((((.....)))).)).)..)))).)).((((..((((((.(((..-...))).)))))))))).((((((((.(.(.(((....))).).).))))))))..... ( -22.10) >DroPse_CAF1 19888 108 + 1 UCUGCACUCCUCUUG------------UUGUGUCGAGGAGCACAAAACACUUUUCUGUUUUUUAACGCGAAAGUUUUGUUAUGGAAAUGCGGCAAGUUUUUGCUACUUUUCCAUAUUUCU ......((((((...------------.......)))))).(((((((...((((((((....)))).)))))))))))((((((((.(..((((....))))..).))))))))..... ( -29.10) >DroSim_CAF1 16534 119 + 1 UUUGCACUUUUCCUGGCGUUUUUUGCCUAGUGUCGAGUAGCUCAAAACACUUUUCUGUUU-UUAACGCAAAAGUUUUGUUAUGGAAAUGCAGCAAGUUCUUGGUACUUUUUCAUAUUUUU ...((((((..(((((((.....)))).)).)..)))).)).((((..((((((.(((..-...))).)))))))))).((((((((.(.(.(((....))).).).))))))))..... ( -22.10) >DroPer_CAF1 20213 108 + 1 UCUGCACUCCUCUUG------------UUGUGUCGAGGAGCACAAAACACUUUUCUGUUUUUUAACGCGAAAGUUUUGUUAUGGAAAUGCGGCAAGUUUUUGCUACUUUUCCAUAUUUCU ......((((((...------------.......)))))).(((((((...((((((((....)))).)))))))))))((((((((.(..((((....))))..).))))))))..... ( -29.10) >consensus UCUGCACUCCUCCUG____________UAGUGUCGAGGAGCACAAAACACUUUUCUGUUU_UUAACGCAAAAGUUUUGUUAUGGAAAUGCAGCAAGUUCUUGCUACUUUUCCAUAUUUCU ......((((((......................))))))..((((..((((((.((((....)))).)))))))))).((((((((.(.(((((....))))).).))))))))..... (-18.90 = -18.90 + 0.00)


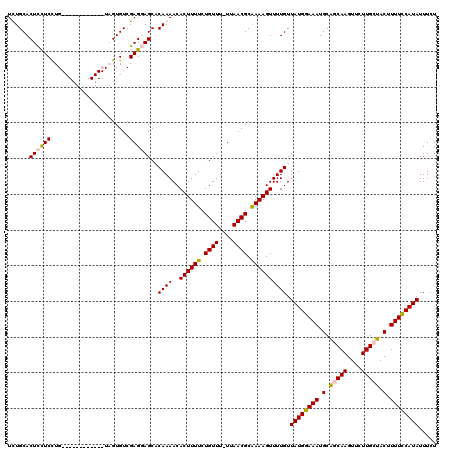
| Location | 13,651,303 – 13,651,422 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.29 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -21.05 |
| Energy contribution | -24.30 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13651303 119 - 22407834 AAAAAUAUGAAAAAGUACCAAGAACUUGCUGCAUUUCCAUAACAAAACUUUUGCGUUAA-AAACAGAAAAGUGUUUUGAGCUACUCGACACUAGGCAAAAAACGCCAGGAAAAGUGCAAA .....((((.(((.(((.(((....))).))).))).)))).....((((((.(.....-.........((((((..(((...))))))))).(((.......))).).))))))..... ( -21.60) >DroPse_CAF1 19888 108 - 1 AGAAAUAUGGAAAAGUAGCAAAAACUUGCCGCAUUUCCAUAACAAAACUUUCGCGUUAAAAAACAGAAAAGUGUUUUGUGCUCCUCGACACAA------------CAAGAGGAGUGCAGA .....((((((((.(..((((....))))..).)))))))).....((((((..(((....))).).)))))..((((..((((((.......------------...))))))..)))) ( -33.90) >DroSim_CAF1 16534 119 - 1 AAAAAUAUGAAAAAGUACCAAGAACUUGCUGCAUUUCCAUAACAAAACUUUUGCGUUAA-AAACAGAAAAGUGUUUUGAGCUACUCGACACUAGGCAAAAAACGCCAGGAAAAGUGCAAA .....((((.(((.(((.(((....))).))).))).)))).....((((((.(.....-.........((((((..(((...))))))))).(((.......))).).))))))..... ( -21.60) >DroPer_CAF1 20213 108 - 1 AGAAAUAUGGAAAAGUAGCAAAAACUUGCCGCAUUUCCAUAACAAAACUUUCGCGUUAAAAAACAGAAAAGUGUUUUGUGCUCCUCGACACAA------------CAAGAGGAGUGCAGA .....((((((((.(..((((....))))..).)))))))).....((((((..(((....))).).)))))..((((..((((((.......------------...))))))..)))) ( -33.90) >consensus AAAAAUAUGAAAAAGUACCAAAAACUUGCCGCAUUUCCAUAACAAAACUUUCGCGUUAA_AAACAGAAAAGUGUUUUGAGCUACUCGACACAA____________CAAGAAAAGUGCAAA .....((((((((.(((((((....))))))).)))))))).....((((((..(((....)))..))))))..((((((((((((......................)))))))))))) (-21.05 = -24.30 + 3.25)


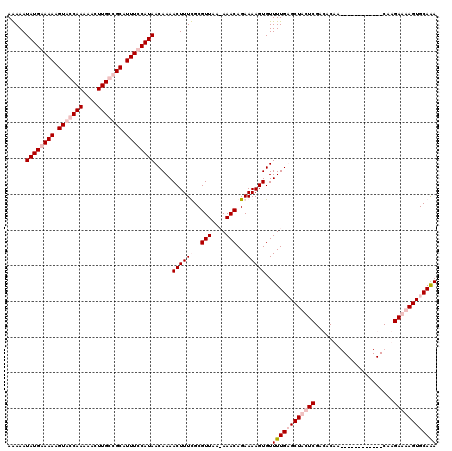
| Location | 13,651,343 – 13,651,462 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -17.64 |
| Consensus MFE | -15.85 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13651343 119 + 22407834 CUCAAAACACUUUUCUGUUU-UUAACGCAAAAGUUUUGUUAUGGAAAUGCAGCAAGUUCUUGGUACUUUUUCAUAUUUUUCUUCGUUCCAUGUCGCUACUUUUUUUGUAUCUAUUUCAUU ...((((((......)))))-)..(((.((((((..((.(((((((.((.((.(((....(((.......)))....))))).))))))))).))..))))))..)))............ ( -15.20) >DroPse_CAF1 19916 111 + 1 CACAAAACACUUUUCUGUUUUUUAACGCGAAAGUUUUGUUAUGGAAAUGCGGCAAGUUUUUGCUACUUUUCCAUAUUUCUGCUAGAUUCCUUUUUU--UUUUUUUUGCCACCA------- .(((((((...((((((((....)))).)))))))))))((((((((.(..((((....))))..).)))))))).....................--...............------- ( -21.00) >DroSim_CAF1 16574 119 + 1 CUCAAAACACUUUUCUGUUU-UUAACGCAAAAGUUUUGUUAUGGAAAUGCAGCAAGUUCUUGGUACUUUUUCAUAUUUUUCUUCGCUUCAUGUCGCUACUUUUUUUGUAUCUAUUUCAUU ...((((((......)))))-)....(((((((......((((((((.(.(.(((....))).).).)))))))).........((........)).....)))))))............ ( -13.30) >DroPer_CAF1 20241 111 + 1 CACAAAACACUUUUCUGUUUUUUAACGCGAAAGUUUUGUUAUGGAAAUGCGGCAAGUUUUUGCUACUUUUCCAUAUUUCUGCUGGAUUCCUUUUAU--UUUUUUUUGCCACCA------- .(((((((...((((((((....)))).)))))))))))((((((((.(..((((....))))..).)))))))).....(.(((...........--.........))).).------- ( -21.05) >consensus CACAAAACACUUUUCUGUUU_UUAACGCAAAAGUUUUGUUAUGGAAAUGCAGCAAGUUCUUGCUACUUUUCCAUAUUUCUCCUCGAUUCAUGUCGC__CUUUUUUUGCAACCA_______ ..(((((.((((((.((((....)))).)))))).....((((((((.(.(((((....))))).).))))))))...........................)))))............. (-15.85 = -15.60 + -0.25)

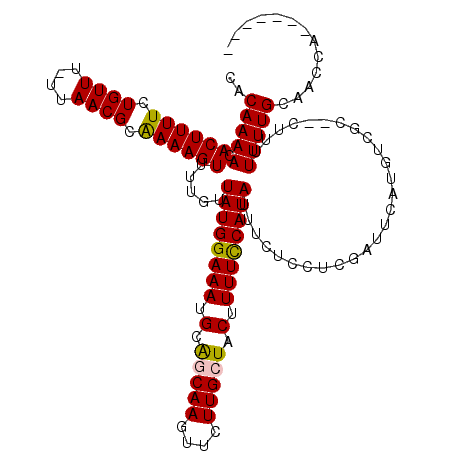

| Location | 13,651,343 – 13,651,462 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.90 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13651343 119 - 22407834 AAUGAAAUAGAUACAAAAAAAGUAGCGACAUGGAACGAAGAAAAAUAUGAAAAAGUACCAAGAACUUGCUGCAUUUCCAUAACAAAACUUUUGCGUUAA-AAACAGAAAAGUGUUUUGAG .............(((((...((((((((.(((.((..................)).))).)...)))))))..............((((((..((...-..))..)))))).))))).. ( -15.37) >DroPse_CAF1 19916 111 - 1 -------UGGUGGCAAAAAAAA--AAAAAAGGAAUCUAGCAGAAAUAUGGAAAAGUAGCAAAAACUUGCCGCAUUUCCAUAACAAAACUUUCGCGUUAAAAAACAGAAAAGUGUUUUGUG -------...............--.....................((((((((.(..((((....))))..).))))))))(((((((((((..(((....))).))))...))))))). ( -22.30) >DroSim_CAF1 16574 119 - 1 AAUGAAAUAGAUACAAAAAAAGUAGCGACAUGAAGCGAAGAAAAAUAUGAAAAAGUACCAAGAACUUGCUGCAUUUCCAUAACAAAACUUUUGCGUUAA-AAACAGAAAAGUGUUUUGAG ........((((((...((((((.((........)).........((((.(((.(((.(((....))).))).))).)))).....))))))..((...-..))......)))))).... ( -13.50) >DroPer_CAF1 20241 111 - 1 -------UGGUGGCAAAAAAAA--AUAAAAGGAAUCCAGCAGAAAUAUGGAAAAGUAGCAAAAACUUGCCGCAUUUCCAUAACAAAACUUUCGCGUUAAAAAACAGAAAAGUGUUUUGUG -------((.(((.........--...........))).))....((((((((.(..((((....))))..).))))))))(((((((((((..(((....))).))))...))))))). ( -24.35) >consensus _______UAGAGACAAAAAAAA__ACAAAAGGAAUCGAACAAAAAUAUGAAAAAGUACCAAAAACUUGCCGCAUUUCCAUAACAAAACUUUCGCGUUAA_AAACAGAAAAGUGUUUUGAG .............................................((((((((.(((((((....))))))).)))))))).((((((((((..(((....))).))))...)))))).. (-15.65 = -16.90 + 1.25)


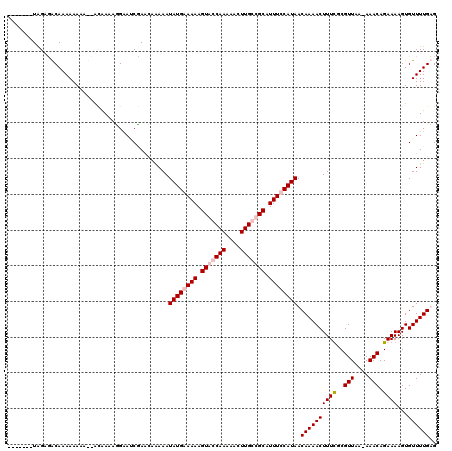
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:10 2006