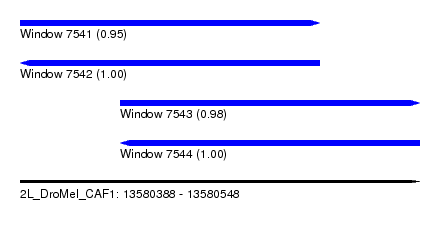
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,580,388 – 13,580,548 |
| Length | 160 |
| Max. P | 0.999945 |

| Location | 13,580,388 – 13,580,508 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -36.66 |
| Consensus MFE | -34.08 |
| Energy contribution | -34.12 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13580388 120 + 22407834 UAUGUGUAUGGGAGCCAUGUCAGUGCCAUUGCAGUUACAUUUUGCUUUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCGUGGCCUGGUCGUUGCCAUUGGC .....(.((.((.(((((((((((.....((((....((((.((((((...(((....)))))))))....)))).......)))).....))))))))))))).)))...(((...))) ( -37.50) >DroSec_CAF1 13493 120 + 1 UGUAUGUAUGGGAGCCAUGGCAGUGCCAUUGCAGUUACAUUUUGCUUUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCGUGGCCUGGUCGUUGCCAUUGGC .((....((((.(((((.(((..(((((..((((........((((((...(((....)))))))))....(((((.......))))).)))))))))..)))))))...).))))..)) ( -37.10) >DroSim_CAF1 12207 116 + 1 ----UGUAUGGGAGUCAUGGCAGUGCCAUUGCAGUUACAUUUUGCUUUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCGUGGCCUGGUCGUUGCCAUUGGC ----.........(((((((((((((((..((((........((((((...(((....)))))))))....(((((.......))))).)))).(((...)))))).)))))))).)))) ( -33.70) >DroEre_CAF1 13842 120 + 1 CACGUGUAUGGAAGCCAUGGCAGUGCCAUUGCAGUUACAUUUUGCUGUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCCUGGCCUGGUCGUUGCCAUUGGC .............((((((((((..((((.(((((........))))).))))..........((((....(((((.......)))))..))))((((......)))).)))))).)))) ( -38.10) >DroYak_CAF1 10634 120 + 1 CACGUGUAAGGGAGCCAUGGCAGUGCCAUUGCAGUUACAAUUUGCUUUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCCUGGCCUGGUCGUUGCCAUUGGC ...(.((((.(..((((.(((((.((((..((((....((..((((((...(((....)))))))))..))(((((.......))))).)))))))))).)))))))).)))))...... ( -36.90) >consensus CACGUGUAUGGGAGCCAUGGCAGUGCCAUUGCAGUUACAUUUUGCUUUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCGUGGCCUGGUCGUUGCCAUUGGC .............(((((((((((((((..((((.......)))).....((((....)))).((((....(((((.......)))))..))))(((...)))))).)))))))).)))) (-34.08 = -34.12 + 0.04)

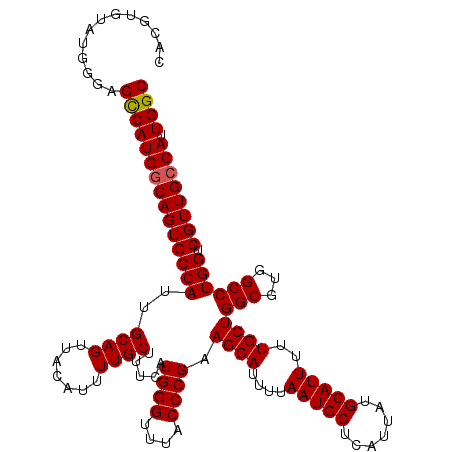
| Location | 13,580,388 – 13,580,508 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -31.42 |
| Energy contribution | -31.82 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.74 |
| SVM RNA-class probability | 0.999945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13580388 120 - 22407834 GCCAAUGGCAACGACCAGGCCACGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGAAAGCAAAAUGUAACUGCAAUGGCACUGACAUGGCUCCCAUACACAUA ((((.((((.........)))).(((((((....(((((...((((((....)))))).(((....)))...........)))))..)))..)))).......))))............. ( -30.70) >DroSec_CAF1 13493 120 - 1 GCCAAUGGCAACGACCAGGCCACGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGAAAGCAAAAUGUAACUGCAAUGGCACUGCCAUGGCUCCCAUACAUACA ((((.(((((..(.((((((...))).(((....(((((...((((((....)))))).(((....)))...........)))))..)))..))))..)))))))))............. ( -34.90) >DroSim_CAF1 12207 116 - 1 GCCAAUGGCAACGACCAGGCCACGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGAAAGCAAAAUGUAACUGCAAUGGCACUGCCAUGACUCCCAUACA---- ....((((((..(.((((((...))).(((....(((((...((((((....)))))).(((....)))...........)))))..)))..))))..))))))............---- ( -29.20) >DroEre_CAF1 13842 120 - 1 GCCAAUGGCAACGACCAGGCCAGGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGACAGCAAAAUGUAACUGCAAUGGCACUGCCAUGGCUUCCAUACACGUG ((((.(((((..(.((((((...))).(((....(((((...((((((....)))))).(((....)))...........)))))..)))..))))..)))))))))............. ( -34.90) >DroYak_CAF1 10634 120 - 1 GCCAAUGGCAACGACCAGGCCAGGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGAAAGCAAAUUGUAACUGCAAUGGCACUGCCAUGGCUCCCUUACACGUG ((((.(((((..(.(((...(((....((((...(((.....((((((....)))))).(((....)))......)))..))))..)))...))))..)))))))))............. ( -35.00) >consensus GCCAAUGGCAACGACCAGGCCACGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGAAAGCAAAAUGUAACUGCAAUGGCACUGCCAUGGCUCCCAUACACAUA ((((.(((((..(.((((((...))).(((..(((((.......)))))....(((((((((....)))..)).)))).........)))..))))..)))))))))............. (-31.42 = -31.82 + 0.40)

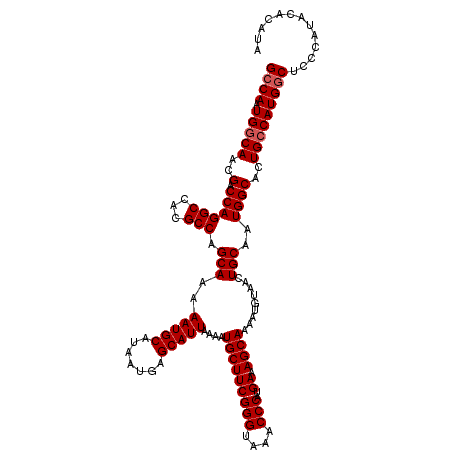
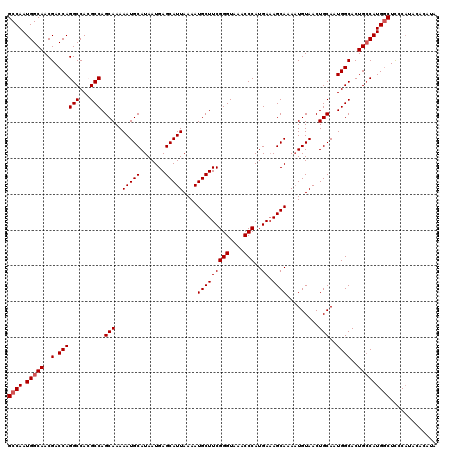
| Location | 13,580,428 – 13,580,548 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.34 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13580428 120 + 22407834 UUUGCUUUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCGUGGCCUGGUCGUUGCCAUUGGCAUUACAACUGCCGCCGAAUCCACCCACUCCUGUUUUAUUA ..........(((((((......((((....(((((.......)))))..))))((((((((.........))))).((((.......)))))))))))))).................. ( -30.60) >DroSec_CAF1 13533 120 + 1 UUUGCUUUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCGUGGCCUGGUCGUUGCCAUUGGCAUUACAACUGCCGCCGAAUCCACCCACUCCUGUUUUAUUA ..........(((((((......((((....(((((.......)))))..))))((((((((.........))))).((((.......)))))))))))))).................. ( -30.60) >DroSim_CAF1 12243 120 + 1 UUUGCUUUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCGUGGCCUGGUCGUUGCCAUUGGCAUUACAACUGCCGCCGAAUCCACCCACUCCUGCUUUAUUA ...((.....(((((........((((....(((((.......)))))..))))((((((((.........))))).((((.......)))))))......))))).....))....... ( -32.20) >DroEre_CAF1 13882 120 + 1 UUUGCUGUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCCUGGCCUGGUCGUUGCCAUUGGCAUUACAACUGCCGCCGCGUCCACCCACUCCUGUUUUAUUA ..........((((....)))).((((....(((((.......)))))..))))((..(((..(((.(((.((....((((.......)))))).))).))).)))..)).......... ( -31.50) >DroYak_CAF1 10674 120 + 1 UUUGCUUUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCCUGGCCUGGUCGUUGCCAUUGGCAUUACAACGGCCGCCGAAUCCACCCACUCCUGUUUUAUUA ..........(((((........((((....(((((.......)))))..))))((..((((..((((((((((...)))).....))))))))))...))))))).............. ( -32.50) >consensus UUUGCUUUCAUGGGUUUACCCGAAGCAUUUUAAUGCUCAUUAUGCAUUUUUGCUGGCGUGGCCUGGUCGUUGCCAUUGGCAUUACAACUGCCGCCGAAUCCACCCACUCCUGUUUUAUUA ...((.....(((((........((((....(((((.......)))))..))))((..((((.((((....))))..((((.......))))))))...))))))).....))....... (-29.30 = -29.34 + 0.04)

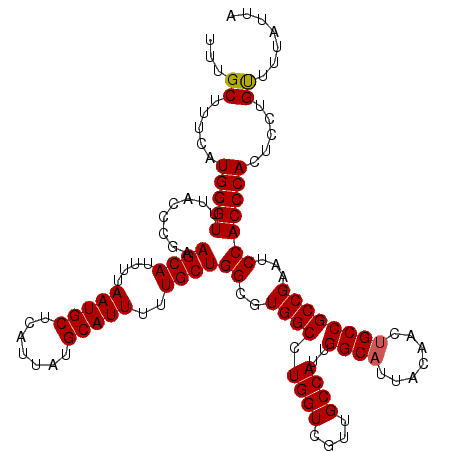
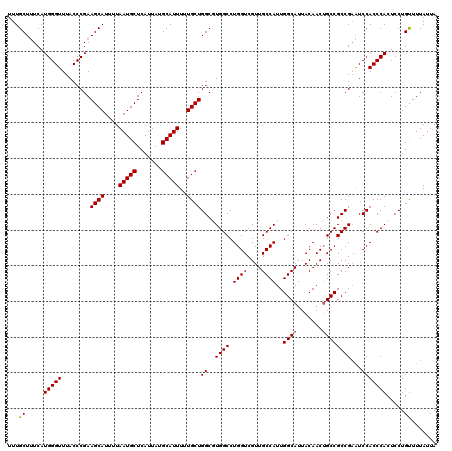
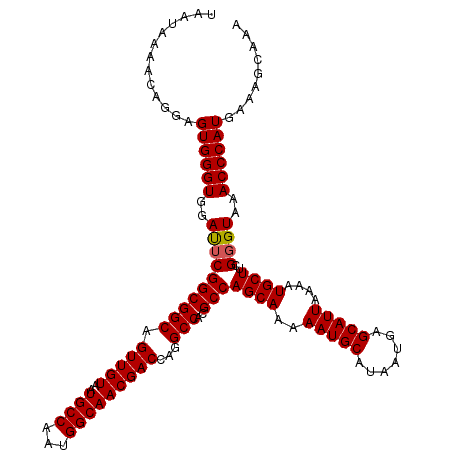
| Location | 13,580,428 – 13,580,548 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -38.06 |
| Consensus MFE | -36.00 |
| Energy contribution | -36.04 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.43 |
| SVM RNA-class probability | 0.999897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13580428 120 - 22407834 UAAUAAAACAGGAGUGGGUGGAUUCGGCGGCAGUUGUAAUGCCAAUGGCAACGACCAGGCCACGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGAAAGCAAA .............((((((..((((((((((.(((((..((((...)))))))))...)))..)))((((..(((((.......)))))....))))..))))..))))))......... ( -36.50) >DroSec_CAF1 13533 120 - 1 UAAUAAAACAGGAGUGGGUGGAUUCGGCGGCAGUUGUAAUGCCAAUGGCAACGACCAGGCCACGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGAAAGCAAA .............((((((..((((((((((.(((((..((((...)))))))))...)))..)))((((..(((((.......)))))....))))..))))..))))))......... ( -36.50) >DroSim_CAF1 12243 120 - 1 UAAUAAAGCAGGAGUGGGUGGAUUCGGCGGCAGUUGUAAUGCCAAUGGCAACGACCAGGCCACGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGAAAGCAAA .......((....((((((..((((((((((.(((((..((((...)))))))))...)))..)))((((..(((((.......)))))....))))..))))..))))))....))... ( -39.40) >DroEre_CAF1 13882 120 - 1 UAAUAAAACAGGAGUGGGUGGACGCGGCGGCAGUUGUAAUGCCAAUGGCAACGACCAGGCCAGGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGACAGCAAA ..........(..((((((..((.(((((((.(((((..((((...)))))))))...)))..)))((((..(((((.......)))))....))))..).))..))))))..)...... ( -39.30) >DroYak_CAF1 10674 120 - 1 UAAUAAAACAGGAGUGGGUGGAUUCGGCGGCCGUUGUAAUGCCAAUGGCAACGACCAGGCCAGGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGAAAGCAAA .............((((((..((((((((((((((((..((((...)))))))))..))))..)))((((..(((((.......)))))....))))..))))..))))))......... ( -38.60) >consensus UAAUAAAACAGGAGUGGGUGGAUUCGGCGGCAGUUGUAAUGCCAAUGGCAACGACCAGGCCACGCCAGCAAAAAUGCAUAAUGAGCAUUAAAAUGCUUCGGGUAAACCCAUGAAAGCAAA .............((((((..((((((((((.(((((..((((...)))))))))...)))..)))((((..(((((.......)))))....))))..))))..))))))......... (-36.00 = -36.04 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:48 2006