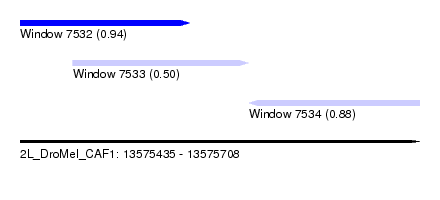
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,575,435 – 13,575,708 |
| Length | 273 |
| Max. P | 0.941484 |

| Location | 13,575,435 – 13,575,551 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -29.77 |
| Energy contribution | -30.77 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13575435 116 + 22407834 ----CAAUGACAUUUUUGAAGCAAACUCCUUUAGAUUAUCUAUUGCAUUCAUCCAGAUAACUUCGGGCGCACAGUGCAUCGGGGAAAAUGGUUGCACUGGGAUGGUUGCAGUGGACUUCC ----..........(((((((.......)))))))...(((((((((..(((((......(....).....((((((((((.......))).))))))))))))..)))))))))..... ( -35.90) >DroSec_CAF1 8604 116 + 1 ----CAAUGACAUUUUUAAAGCAAACUCCUUUAGAUUAUCUAUUGCAUUCAUCCAGAUAACUACGAGCGCACAGUGCAUCGUGGAAAAUGGUUGCACUGGGAUGGUUGCAGUGGACUUCC ----..........(((((((.......)))))))...(((((((((..(((((((....)).........(((((((((((.....)))).))))))))))))..)))))))))..... ( -35.10) >DroSim_CAF1 8380 116 + 1 ----AAAUGACAUUUUUGAAGCAAACUCCUUUAGAUUAUCUAUUGCAUUCAUCCAGAUAACUACGAGCGCACAGUGCAUCGUGGAAAAUGGUUGCACUGGGAUGGUUGCAGUGGACUUCC ----..........(((((((.......)))))))...(((((((((..(((((((....)).........(((((((((((.....)))).))))))))))))..)))))))))..... ( -35.20) >DroEre_CAF1 9086 117 + 1 UUUACAAUCACAUCUUCGAAACAAACUCCUUUAGAUAAUCUAUGGUAUGCAUCUUGAUGACUUUGGGAACACAGUGCACCGUUGAAAAUGGUUGCACUGGGAU---GGCAGUGGACUUCC ....((....(((((...........((((..((...(((...(((....)))..)))..))..))))...(((((((((((.....)))).)))))))))))---)....))....... ( -28.70) >consensus ____CAAUGACAUUUUUGAAGCAAACUCCUUUAGAUUAUCUAUUGCAUUCAUCCAGAUAACUACGAGCGCACAGUGCAUCGUGGAAAAUGGUUGCACUGGGAUGGUUGCAGUGGACUUCC ..............(((((((.......)))))))...(((((((((.((((((..........(....).(((((((((((.....)))).))))))))))))).)))))))))..... (-29.77 = -30.77 + 1.00)

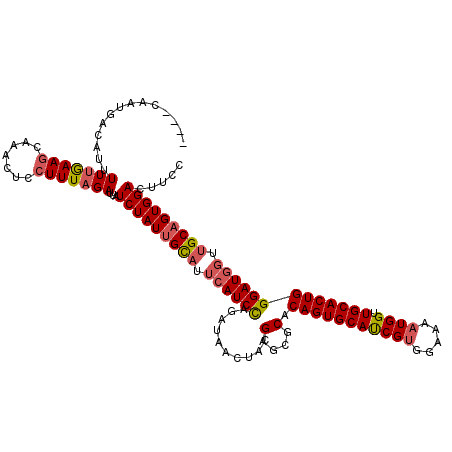
| Location | 13,575,471 – 13,575,591 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -28.80 |
| Energy contribution | -29.16 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13575471 120 + 22407834 UAUUGCAUUCAUCCAGAUAACUUCGGGCGCACAGUGCAUCGGGGAAAAUGGUUGCACUGGGAUGGUUGCAGUGGACUUCCGUUAUGGUCAUAGCAACCUGCUCAACAUGUUAAUGCAACG ..((((((((((...((.....))((((...((((((((((.......))).)))))))....((((((.(((.(((........)))))).)))))).))))...)))..))))))).. ( -36.80) >DroSec_CAF1 8640 120 + 1 UAUUGCAUUCAUCCAGAUAACUACGAGCGCACAGUGCAUCGUGGAAAAUGGUUGCACUGGGAUGGUUGCAGUGGACUUCCGUUAUGGUCAUAGCAACCUGCUCAACAUGUUAAUGCAACG ..((((((((.....))((((...((((...(((((((((((.....)))).)))))))....((((((.(((.(((........)))))).)))))).)))).....)))))))))).. ( -38.30) >DroSim_CAF1 8416 120 + 1 UAUUGCAUUCAUCCAGAUAACUACGAGCGCACAGUGCAUCGUGGAAAAUGGUUGCACUGGGAUGGUUGCAGUGGACUUCCGUUAUGGUCAUAGCAACCUGCUCAACAUGUUAAUGCAACG ..((((((((.....))((((...((((...(((((((((((.....)))).)))))))....((((((.(((.(((........)))))).)))))).)))).....)))))))))).. ( -38.30) >DroEre_CAF1 9126 117 + 1 UAUGGUAUGCAUCUUGAUGACUUUGGGAACACAGUGCACCGUUGAAAAUGGUUGCACUGGGAU---GGCAGUGGACUUCCGUUAUGGUCAUAGCAACCUGCUCAACAUGUUAUUGCAACG ....(((.((((.(((((((((.((....))(((((((((((.....)))).))))))).(((---((.((....)).)))))..))))))(((.....)))))).))))...))).... ( -33.80) >DroYak_CAF1 5774 105 + 1 ---------------GAUAACUUUGGGAACACAGUGCAUCGUUGAAAAUGGUUGCACUGGGAUGGUGGCAGUGGACUUCCGUUAUGGUCAUAGCAACCUGCUCAACAUGUUAAUGCAACG ---------------(((.(((.((....)).)))..)))..........((((((.(((.(((.((((((.((....))(((((....)))))...)))).)).))).))).)))))). ( -30.90) >consensus UAUUGCAUUCAUCCAGAUAACUUCGGGCGCACAGUGCAUCGUGGAAAAUGGUUGCACUGGGAUGGUUGCAGUGGACUUCCGUUAUGGUCAUAGCAACCUGCUCAACAUGUUAAUGCAACG ....................((((((((((...)))).))))))......((((((.(((.(((...((((.((....))(((((....)))))...))))....))).))).)))))). (-28.80 = -29.16 + 0.36)



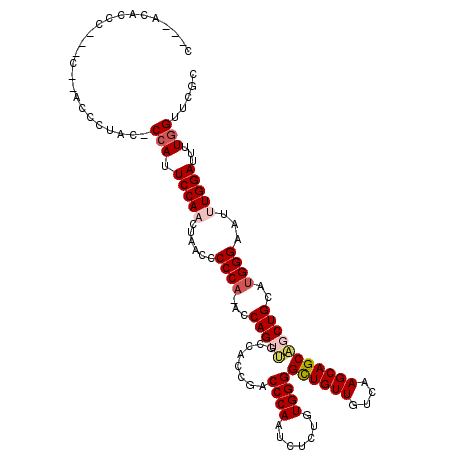
| Location | 13,575,591 – 13,575,708 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -24.42 |
| Energy contribution | -25.34 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13575591 117 - 22407834 C---ACACCCAACCGCACCCUACCCGAUUCCAACUAACCCCCAAACCAGUUGCCACCGACCCAAUCUCUGUGGGGCUGUUGUCAAGCAGCAGCUGCAUGGGAAUUUGGAUUUUGGUUCGC .---..........((.....(((.((.(((((......((((...(((((((......((((.......))))(((.......))).)))))))..))))...))))).)).)))..)) ( -31.10) >DroSec_CAF1 8760 116 - 1 CUCCACACCC---CACACCCUACACCAUUCCAACUAAACCCCA-ACCAGUUGCCACCGACCCAAUCCCUGUGGGGCUGUUGUCAAGCAGCAGCUGCAUGGGAAUUUGGAUUUUGGUUCGC ..........---..........((((.(((((..........-....((((....))))....((((.(((.((((((((.....)))))))).)))))))..)))))...)))).... ( -33.30) >DroSim_CAF1 8536 114 - 1 CUCCACACCC---C--ACCCUACCCCAUUCCAACUAAACCCCA-ACCAGUUGCCACCGACCCAAUCCCUGUGGGGCUGUUGUCAAGCAGCAGCUGCAUGGGAAUUUGGAUUUUGGUUCGC ..........---.--...............(((((((..(((-(...((((....))))....((((.(((.((((((((.....)))))))).)))))))..))))..)))))))... ( -32.50) >DroEre_CAF1 9243 94 - 1 ------------------------CCAUUCCACCUAACCCCCA-ACCAGA-UCCACCGACCCAAUCUCCGUGGGGUUGUUGUCAAGCAGCUGCUGCAUGGGAAUUUGGAUUUUGGUUCGC ------------------------..................(-((((((-((((.......(((..(((((.(((.((((.....)))).))).)))))..))))))).)))))))... ( -27.41) >DroYak_CAF1 5879 93 - 1 ------------------------CCAUUCCACCCAACCCCCA-ACCAGAACCCAACGCCCCAAUCUCUGUGGGGUUGUUGUCAAGCAGCUGCUGC--GGGAAUUUGGAUUUUGGUUCGC ------------------------(((.(((((((........-.........((((((((((.......))))).)))))....((((...))))--)))....))))...)))..... ( -27.40) >consensus C___ACACCC___C__ACCCUAC_CCAUUCCAACUAACCCCCA_ACCAGUUGCCACCGACCCAAUCUCUGUGGGGCUGUUGUCAAGCAGCAGCUGCAUGGGAAUUUGGAUUUUGGUUCGC ........................(((.(((((......((((...(((((........((((.......))))((((((....)))))))))))..))))...)))))...)))..... (-24.42 = -25.34 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:37 2006