| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,575,059 – 13,575,174 |
| Length | 115 |
| Max. P | 0.878981 |

| Location | 13,575,059 – 13,575,174 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.77 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -17.46 |
| Energy contribution | -19.06 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13575059 115 + 22407834 CUCCGUCCUGCCACCAACUGCCACCCACAGCCCAACACCCCAUCAUC-----CCACCCAUUACCCAUUACCCACGGGCGCAUUGUUUACUGUGGUGUGCAGCAUAUUGGUUGAAGUAAAA .......((.(.(((((((((((((.((((...((((((((......-----......................))).)...))))..)))))))).))))....))))).).))..... ( -23.09) >DroSec_CAF1 8269 120 + 1 CUUCGUCCUUCCACCAACAGCCACCCACAGCCCAACUCCCCAUCACCCCAUCUCACCCAUUACCCAUUACCCACGGGCUCAUUGUUUACUGUGGUGUGCAGCAUAUUGGUUGAAGUAAAA .......((((.(((((..(((((((((((..(((...(((.................................)))....)))....)))))).)))..))...))))).))))..... ( -23.81) >DroSim_CAF1 8056 120 + 1 NNNNNNNNNNNNACCAACUGCCACCCACCGCCCAACUCCCCAUCACCCCAUCUCACCCAUUACCCAUUACCCACGGGCUCAUUGUUUACUGUGGUGUGCAGCAUAUUGGUUGAAGUAAAA ............(((((((((((((((..((((.........................................)))).....(....))).)))).))))....))))).......... ( -18.28) >DroEre_CAF1 8732 116 + 1 CUCCGCCCUUCCACCAGCUGCCACCCACAGCCCAACAC-CCAUUACC-CA--UUACCCAUUACCCAUUACCCACGGGCGCUUUGUUUACUGUGGUGUGCAGCAUAUUGGCUGGAGUAAUA (((((((.........(((((((((.((((...((((.-......((-(.--......................))).....))))..)))))))).))))).....))).))))..... ( -30.14) >DroYak_CAF1 5404 116 + 1 CUCCGCCCUUCCACCAACUGCCACCCACAGCCCACAGC-CCAGAUCC-CA--UCACCAAGCACCCAUUACCCACGGGUGCAUUGUUUACUGUGGUGUGAAGACUAUUGGUUGACGUUAUA .........((.(((((((..(((((((((...((((.-...(((..-.)--)).....((((((.........)))))).))))...)))))).))).))....))))).))....... ( -30.60) >consensus CUCCGCCCUUCCACCAACUGCCACCCACAGCCCAACACCCCAUCACC_CA__UCACCCAUUACCCAUUACCCACGGGCGCAUUGUUUACUGUGGUGUGCAGCAUAUUGGUUGAAGUAAAA .......((((.(((((((((((((.(((((((.........................................)))).....(....)))))))).))))....))))).))))..... (-17.46 = -19.06 + 1.60)

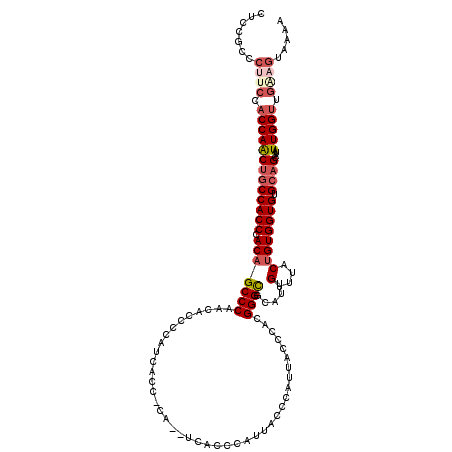
| Location | 13,575,059 – 13,575,174 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.77 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -28.56 |
| Energy contribution | -30.24 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13575059 115 - 22407834 UUUUACUUCAACCAAUAUGCUGCACACCACAGUAAACAAUGCGCCCGUGGGUAAUGGGUAAUGGGUGG-----GAUGAUGGGGUGUUGGGCUGUGGGUGGCAGUUGGUGGCAGGACGGAG .....((((.((((((.(((..(...(((((((...((((((.(((((.(..................-----..).))))))))))).))))))))..)))))))))(......))))) ( -38.05) >DroSec_CAF1 8269 120 - 1 UUUUACUUCAACCAAUAUGCUGCACACCACAGUAAACAAUGAGCCCGUGGGUAAUGGGUAAUGGGUGAGAUGGGGUGAUGGGGAGUUGGGCUGUGGGUGGCUGUUGGUGGAAGGACGAAG .....((((.(((((((.((..(...(((((((...((((...(((((.(.(......((.....))......).).)))))..)))).))))))))..))))))))).))))....... ( -39.50) >DroSim_CAF1 8056 120 - 1 UUUUACUUCAACCAAUAUGCUGCACACCACAGUAAACAAUGAGCCCGUGGGUAAUGGGUAAUGGGUGAGAUGGGGUGAUGGGGAGUUGGGCGGUGGGUGGCAGUUGGUNNNNNNNNNNNN ..........((((((.(((..(.((((.((....((.....((((((.....)))))).....))....)).(.(.(........).).))))).)..)))))))))............ ( -29.80) >DroEre_CAF1 8732 116 - 1 UAUUACUCCAGCCAAUAUGCUGCACACCACAGUAAACAAAGCGCCCGUGGGUAAUGGGUAAUGGGUAA--UG-GGUAAUGG-GUGUUGGGCUGUGGGUGGCAGCUGGUGGAAGGGCGGAG .....((((.(((..((((((((...(((((((.((((.(..((((((.....((.....)).....)--))-)))..)..-.))))..)))))))...)))))..)))....))))))) ( -43.70) >DroYak_CAF1 5404 116 - 1 UAUAACGUCAACCAAUAGUCUUCACACCACAGUAAACAAUGCACCCGUGGGUAAUGGGUGCUUGGUGA--UG-GGAUCUGG-GCUGUGGGCUGUGGGUGGCAGUUGGUGGAAGGGCGGAG .....((((.((((((.((((((((((((((((...(((.((((((((.....)))))))))))..((--(.-..)))...-)))))))..)))))).))).)))))).....))))... ( -46.70) >consensus UUUUACUUCAACCAAUAUGCUGCACACCACAGUAAACAAUGCGCCCGUGGGUAAUGGGUAAUGGGUGA__UG_GGUGAUGGGGAGUUGGGCUGUGGGUGGCAGUUGGUGGAAGGACGGAG ..........((((....(((((...(((((((...((((((((((((.....)))))).......................)))))).)))))))...)))))))))............ (-28.56 = -30.24 + 1.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:34 2006