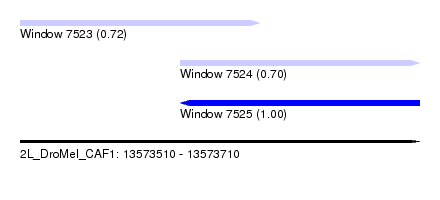
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,573,510 – 13,573,710 |
| Length | 200 |
| Max. P | 0.995565 |

| Location | 13,573,510 – 13,573,630 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -25.63 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13573510 120 + 22407834 AUAUGUAUGUAUGUACAUAGUUACGUAUAGUUAGUUGGCUGAGUGGGGUGCUGACUCUCGUAACUUUGAAGUGUUGCGAACUUGGCGCACUUGAUGGCACUAAUUUAUGCAGGGCCAACA .......((((((((......))))))))....(((((((....(((((((..(...(((((((........)))))))..)..)))).)))....(((........)))..))))))). ( -36.40) >DroSec_CAF1 6751 106 + 1 AUAUGUAU--AUAU-----GUUACGUAUAGUUAGUUGGCU----GGGCUGCUGACUCUCGUA--CUUGAAGU-UUGCGAACUUGGCGCACUUGAUGGCACUAAUUUAUGCCGGGCCAACA (((((((.--....-----..))))))).....(((((((----.((((((.((((.(((..--..))))))-).)))...((((.((.(.....))).)))).....))).))))))). ( -34.30) >DroSim_CAF1 6328 111 + 1 AUUUGUAUAUAUAU-----GUUACGUAUAGUUAGUUGGCU----GGGCUGCUGACUCUCGUAACUUUGAAGUGUUGCGAACUUGGCGCACUUGAUGGCACUAAUUUAUGCAGGGCCAACA .........(((((-----(...))))))....(((((((----..((.((..(...(((((((........)))))))..)..))((.(.....)))..........))..))))))). ( -30.90) >DroEre_CAF1 7205 107 + 1 AUAUGUAG---------UAGUUACGUAUAUUUAGUUGGCU----GGGCUGCUGACUCUCGUAACUUUGAAGUGUUGCGAACUUGGCGCACUUGAUGGCACUAAUUUAUGCAGGGCCAACA ((((((((---------...)))))))).....(((((((----..((.((..(...(((((((........)))))))..)..))((.(.....)))..........))..))))))). ( -32.80) >DroYak_CAF1 3858 104 + 1 AUAUGUA------------GUUACGUAUAGUUAGUUCGCU----GGGGUGCUGACUCUCGUAACUUUGAAGUGUUGCGAACUUGGCGCACUUGAUGGCACUAAUUUAUGCAGGGCCAACA ...(((.------------(((..((((((((((((((..----(((((((..(...(((((((........)))))))..)..)))).)))..))).))))).))))))..)))..))) ( -29.90) >consensus AUAUGUAU__AU_U_____GUUACGUAUAGUUAGUUGGCU____GGGCUGCUGACUCUCGUAACUUUGAAGUGUUGCGAACUUGGCGCACUUGAUGGCACUAAUUUAUGCAGGGCCAACA (((((((..............))))))).....(((((((.........((..(...(((((((........)))))))..)..))(((...(((.......)))..)))..))))))). (-25.63 = -26.28 + 0.65)

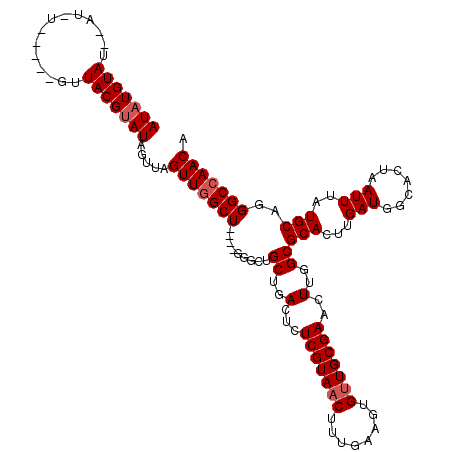
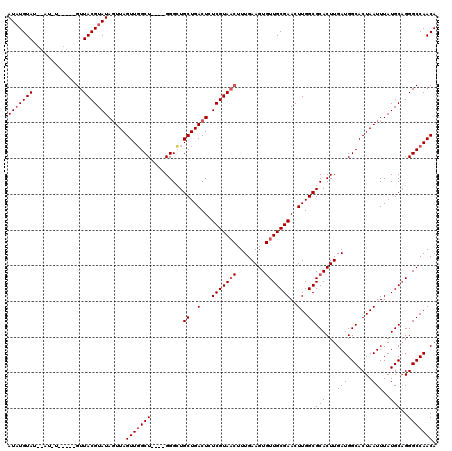
| Location | 13,573,590 – 13,573,710 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -30.56 |
| Energy contribution | -30.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13573590 120 + 22407834 CUUGGCGCACUUGAUGGCACUAAUUUAUGCAGGGCCAACAAUGCCACACCGCAUAAUCAUAAUCCAGACCGCGUGGCUAAUUUGUUUAUAAUUUUCAUAACUUAACGUGUGCGCUAAGCG ((((((((((.((.((((.((..........))))))((((.(((((...((....((........))..)))))))....))))....................)).)))))))))).. ( -30.60) >DroSec_CAF1 6817 120 + 1 CUUGGCGCACUUGAUGGCACUAAUUUAUGCCGGGCCAACAAUGCCACACCGCAUAAUCAUAAUCCAGACCGCGUGGCUAAUUUGUUUAUAAUUUUCAUAACUUAACGUGUGCGCUAAGCG ((((((((((.((.(((((........)))))....(((((.(((((...((....((........))..)))))))....)))))...................)).)))))))))).. ( -31.80) >DroSim_CAF1 6399 120 + 1 CUUGGCGCACUUGAUGGCACUAAUUUAUGCAGGGCCAACAAUGCCACACCGCAUAAUCAUAAUCCAGACCGCGUGGCUAAUUUGUUUAUAAUUUUCAUAACUUAACGUGUGCGCUAAGCG ((((((((((.((.((((.((..........))))))((((.(((((...((....((........))..)))))))....))))....................)).)))))))))).. ( -30.60) >DroEre_CAF1 7272 120 + 1 CUUGGCGCACUUGAUGGCACUAAUUUAUGCAGGGCCAACAAUGCCACACCGCAUAAUCAUAAUCCAGACCGCGUGGCUAAUUUGUUUAUAAUUUUCAUAACUUAACGUGUGCGCUAAGCG ((((((((((.((.((((.((..........))))))((((.(((((...((....((........))..)))))))....))))....................)).)))))))))).. ( -30.60) >DroYak_CAF1 3922 120 + 1 CUUGGCGCACUUGAUGGCACUAAUUUAUGCAGGGCCAACAAUGCCACACCGCAUAAUCAUGAUCCAGACCGCGUGGCUAAUUUGUUUAUAAUUUUCAUAACUUAACGUGUGCGCUAAGCG ((((((((((.((.((((.((..........))))))((((.(((((...((....((........))..)))))))....))))....................)).)))))))))).. ( -30.60) >consensus CUUGGCGCACUUGAUGGCACUAAUUUAUGCAGGGCCAACAAUGCCACACCGCAUAAUCAUAAUCCAGACCGCGUGGCUAAUUUGUUUAUAAUUUUCAUAACUUAACGUGUGCGCUAAGCG ((((((((((.((.((((.((..........))))))((((.(((((...((....((........))..)))))))....))))....................)).)))))))))).. (-30.56 = -30.56 + 0.00)

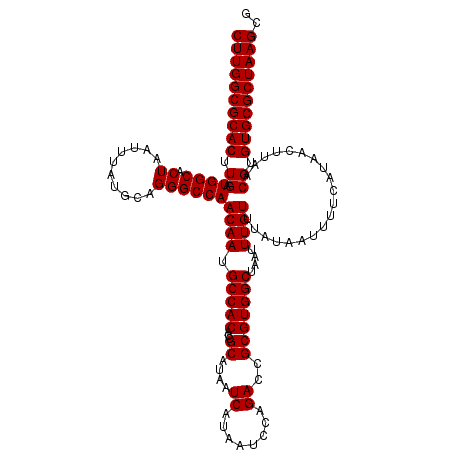
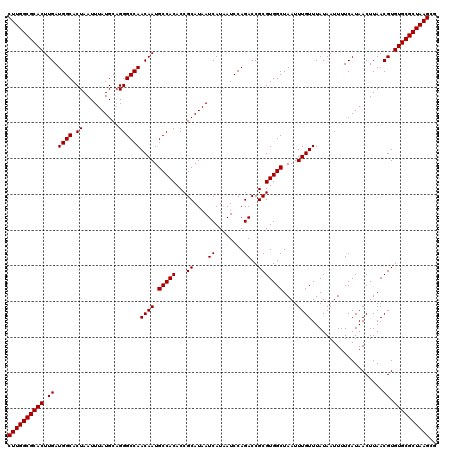
| Location | 13,573,590 – 13,573,710 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -35.92 |
| Energy contribution | -36.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
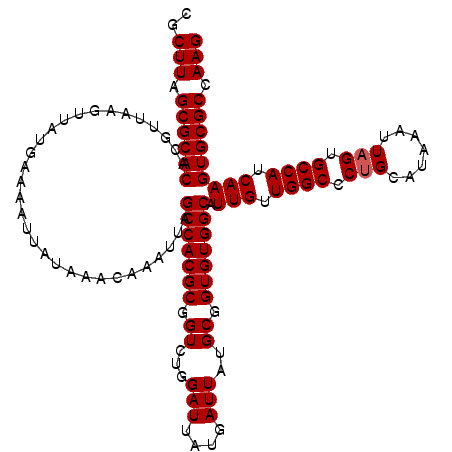
>2L_DroMel_CAF1 13573590 120 - 22407834 CGCUUAGCGCACACGUUAAGUUAUGAAAAUUAUAAACAAAUUAGCCACGCGGUCUGGAUUAUGAUUAUGCGGUGUGGCAUUGUUGGCCCUGCAUAAAUUAGUGCCAUCAAGUGCGCCAAG ..(((.((((((...............................(((((((.((...(((....)))..)).))))))).(((.((((.(((.......))).)))).))))))))).))) ( -36.60) >DroSec_CAF1 6817 120 - 1 CGCUUAGCGCACACGUUAAGUUAUGAAAAUUAUAAACAAAUUAGCCACGCGGUCUGGAUUAUGAUUAUGCGGUGUGGCAUUGUUGGCCCGGCAUAAAUUAGUGCCAUCAAGUGCGCCAAG ..(((.((((((..(((((.((((((...))))))........(((((((.((...(((....)))..)).)))))))....)))))..(((((......))))).....)))))).))) ( -37.40) >DroSim_CAF1 6399 120 - 1 CGCUUAGCGCACACGUUAAGUUAUGAAAAUUAUAAACAAAUUAGCCACGCGGUCUGGAUUAUGAUUAUGCGGUGUGGCAUUGUUGGCCCUGCAUAAAUUAGUGCCAUCAAGUGCGCCAAG ..(((.((((((...............................(((((((.((...(((....)))..)).))))))).(((.((((.(((.......))).)))).))))))))).))) ( -36.60) >DroEre_CAF1 7272 120 - 1 CGCUUAGCGCACACGUUAAGUUAUGAAAAUUAUAAACAAAUUAGCCACGCGGUCUGGAUUAUGAUUAUGCGGUGUGGCAUUGUUGGCCCUGCAUAAAUUAGUGCCAUCAAGUGCGCCAAG ..(((.((((((...............................(((((((.((...(((....)))..)).))))))).(((.((((.(((.......))).)))).))))))))).))) ( -36.60) >DroYak_CAF1 3922 120 - 1 CGCUUAGCGCACACGUUAAGUUAUGAAAAUUAUAAACAAAUUAGCCACGCGGUCUGGAUCAUGAUUAUGCGGUGUGGCAUUGUUGGCCCUGCAUAAAUUAGUGCCAUCAAGUGCGCCAAG ..(((.((((((...............................(((((((.((...(((....)))..)).))))))).(((.((((.(((.......))).)))).))))))))).))) ( -37.30) >consensus CGCUUAGCGCACACGUUAAGUUAUGAAAAUUAUAAACAAAUUAGCCACGCGGUCUGGAUUAUGAUUAUGCGGUGUGGCAUUGUUGGCCCUGCAUAAAUUAGUGCCAUCAAGUGCGCCAAG ..(((.((((((...............................(((((((.((...(((....)))..)).))))))).(((.((((.(((.......))).)))).))))))))).))) (-35.92 = -36.12 + 0.20)

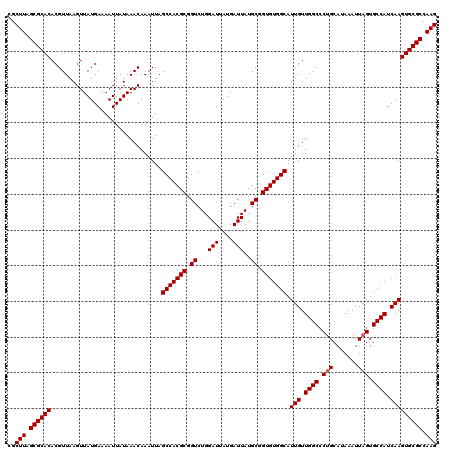
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:28 2006