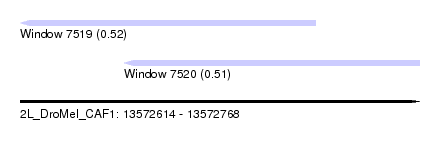
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,572,614 – 13,572,768 |
| Length | 154 |
| Max. P | 0.515147 |

| Location | 13,572,614 – 13,572,728 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -16.21 |
| Energy contribution | -16.97 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13572614 114 - 22407834 UGCUCCCC------AAUGGGGAGGGGGUGGGGGUAAGGCAGAACCCUAGCAAUUCCCCAGAGCGAAUUCAAAACACAAAAGCUCAAUGGACAUGUAGCCCUGUCGAGAUAACAUACACAA ..((((((------...))))))((((((((((....((((....)).))...))))).((((.................))))............)))))................... ( -34.03) >DroSec_CAF1 5889 110 - 1 UGCUCCCCUUCCCCAUAGGGGA----------UAAAGGCAGAACCCUCGCAAUUCCCCAGAGCGAAUUCAAAACACAAAAGCUCAAUGGACAUGUAGCCCUGUCGAGAUAACAUACACAA ...((((((.......))))))----------....(((((....((.(((..(((...((((.................))))...)))..)))))..)))))................ ( -24.13) >DroSim_CAF1 5466 108 - 1 UGCUCCCCUUC--CAUUGGGGA----------UAAAGGCAGAACCCUUGCAAUUCCCCAGAGCGAAUUCAAAACACAAAAGCUCAAUGGACAUGUAGCCCUGUCGAGAUAACAUACACAA .(((.(...((--(((((((((----------.....((((.....))))...))))).((((.................))))))))))...).)))...................... ( -25.93) >DroEre_CAF1 6393 98 - 1 UUAUCCCA-------A-GGGGA-----------GAAGGC---ACCCCACCAAUUCCCCAGAGCGAAUUCACAACACAAAAGCUCAAUGGACAUGUAGCCCUGUCGAGAUAACAUACACAA (((((.(.-------.-(((((-----------(..((.---......))..)))))).((((.................))))....((((........))))).)))))......... ( -21.73) >DroYak_CAF1 2984 101 - 1 UUCUCCCA-------A-UGGGA-----------GAAGGCAGAACCCCACCAAUUCCCCAGAGCGAAUUCAAAACACAAAAGCUCAAUGGACAUGUAGCCCUGUCGAGAUAACAUACACAA (((((((.-------.-.))))-----------)))(((((..(..((.....(((...((((.................))))...)))..))..)..)))))................ ( -21.93) >consensus UGCUCCCC______AA_GGGGA___________AAAGGCAGAACCCUAGCAAUUCCCCAGAGCGAAUUCAAAACACAAAAGCUCAAUGGACAUGUAGCCCUGUCGAGAUAACAUACACAA ...(((((.........)))))..............(((((.......(((..(((...((((.................))))...)))..)))....)))))................ (-16.21 = -16.97 + 0.76)


| Location | 13,572,654 – 13,572,768 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -17.68 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13572654 114 - 22407834 CAGCAUAACAUGCCGCUGGGCUCGGCUCAGAGUUUCCCAGUGCUCCCC------AAUGGGGAGGGGGUGGGGGUAAGGCAGAACCCUAGCAAUUCCCCAGAGCGAAUUCAAAACACAAAA ..((....(((.(((((((((((......)))...)))))).((((((------...)))))))).)))((((....((((....)).))....))))...))................. ( -42.20) >DroSec_CAF1 5929 104 - 1 ------AACAUGCCGCUGGGCUCGGCUCAGAAUUUCUCAGUGCUCCCCUUCCCCAUAGGGGA----------UAAAGGCAGAACCCUCGCAAUUCCCCAGAGCGAAUUCAAAACACAAAA ------....((((.((((((...)))))).............((((((.......))))))----------....))))(((...((((...........)))).)))........... ( -27.20) >DroSim_CAF1 5506 108 - 1 CAGCAUAACAUGCCGCUGGGCUCGGCUCAGAGUUUCCCAGUGCUCCCCUUC--CAUUGGGGA----------UAAAGGCAGAACCCUUGCAAUUCCCCAGAGCGAAUUCAAAACACAAAA ..(((.....(((((((((((((......)))...))))))..(((((...--....)))))----------....)))).......)))(((((........)))))............ ( -31.20) >DroEre_CAF1 6433 98 - 1 CAGCAUAACAUGCCGCUGGGCUCGACUCAGAGUUUCAUAUUUAUCCCA-------A-GGGGA-----------GAAGGC---ACCCCACCAAUUCCCCAGAGCGAAUUCACAACACAAAA ..((((...))))((((((((((......)))))).............-------.-(((((-----------(..((.---......))..))))))..))))................ ( -22.60) >DroYak_CAF1 3024 101 - 1 CAGCAUAACAAGCCGCUGGGCUCGGCUCAGAGUUUCGCAGUUCUCCCA-------A-UGGGA-----------GAAGGCAGAACCCCACCAAUUCCCCAGAGCGAAUUCAAAACACAAAA ..((.......(((((..(((((......)))))..))..(((((((.-------.-.))))-----------))))))..............((....))))................. ( -25.40) >consensus CAGCAUAACAUGCCGCUGGGCUCGGCUCAGAGUUUCCCAGUGCUCCCC______AA_GGGGA___________AAAGGCAGAACCCUAGCAAUUCCCCAGAGCGAAUUCAAAACACAAAA ...........((((((((((((......)))...))))))..(((((.........)))))..............)))...........(((((........)))))............ (-17.68 = -18.20 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:23 2006