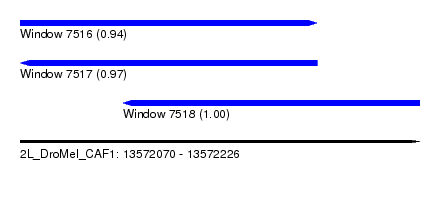
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,572,070 – 13,572,226 |
| Length | 156 |
| Max. P | 0.996107 |

| Location | 13,572,070 – 13,572,186 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.88 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -24.58 |
| Energy contribution | -25.98 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13572070 116 + 22407834 UUGCUUGAUUUGCAUAUGUGAGCGUGUGUAUUCUGCACACAGAAUUUUCUUGUUGCGCUUUUUAUACCCAUUACCCUUGCGCUAAGCGGGUAUAUUAAAUUAUGUGCAAUUUAUG----C ..((.(((.(((((((((.((((((((((.....))))))(((....)))......))))..(((((((.........((.....)))))))))......))))))))).))).)----) ( -31.20) >DroSec_CAF1 5399 116 + 1 UUGCUUGAUUUGCAUAUGUGAGCGCGUGUGUUCUGCACACAGAAUUUUCUUGUUGCGCUUUUUAUACCCAUUACCCUUGGGCUAAGCGGGUAUAUAACAUUUCGAGCAAUUUAUG----C ((((((((..((.......(((((((((((.....)))))(((....)))....))))))..(((((((.(((((....)).)))..)))))))...))..))))))))......----. ( -39.90) >DroSim_CAF1 4943 116 + 1 UUGCUUGAUUUGCAUAUGUGAGCGUGUGUGUUCUGCACACAGAAUUUUCUUGUUGCGCUUUUUAUACCCAUUACCCUUGGACUAAGCGGGUAUAUUAGAUUUCGUGCAAUUUUUG----C ..((..((.((((((....(((((((((((.....)))))(((....)))....))))))..(((((((.(((((...))..)))..))))))).........)))))).))..)----) ( -30.20) >DroEre_CAF1 5851 115 + 1 UUGCUAGAUUUGCAUAUGUGAGCGUGUGUGUUCUGCACACAGAAUUUUCUUGUUGCGCUU-UUAUACCCCUUACCCAUGGGUAAGGCGGGUAUAUCACAUUUCCAGCAUUUUGUG----C ..((..((..(((..(((((((((((((((.....)))))(((....)))....))))..-.(((((((((((((....))))))..))))))))))))).....)))..))..)----) ( -38.50) >DroYak_CAF1 2516 120 + 1 UUGCUUGAUUUGCAUAUGUGAGCGUGUGUGUUCUGCACACAGAAUUUUCUUGUUGCGCUUUUUAUACCCAUUACCCUUGGGCAAAGAGGGUAUCUUACAUUUUGAGCAAUUUAUGUUUGU (((((..(..((..((((.(((((((((((.....)))))(((....)))....))))))..)))).....(((((((.......))))))).....))..)..)))))........... ( -32.50) >consensus UUGCUUGAUUUGCAUAUGUGAGCGUGUGUGUUCUGCACACAGAAUUUUCUUGUUGCGCUUUUUAUACCCAUUACCCUUGGGCUAAGCGGGUAUAUUACAUUUCGAGCAAUUUAUG____C ((((((((...........(((((((((((.....)))))(((....)))....))))))..(((((((....((....))......))))))).......))))))))........... (-24.58 = -25.98 + 1.40)


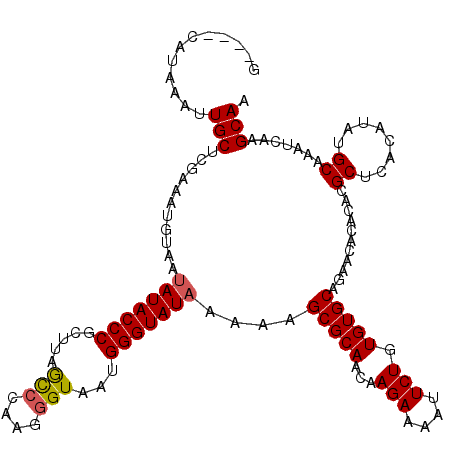
| Location | 13,572,070 – 13,572,186 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.88 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.98 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13572070 116 - 22407834 G----CAUAAAUUGCACAUAAUUUAAUAUACCCGCUUAGCGCAAGGGUAAUGGGUAUAAAAAGCGCAACAAGAAAAUUCUGUGUGCAGAAUACACACGCUCACAUAUGCAAAUCAAGCAA (----((((....((...........((((((((.(((.(.....).)))))))))))....(((((...(((....))).)))))...........)).....)))))........... ( -27.40) >DroSec_CAF1 5399 116 - 1 G----CAUAAAUUGCUCGAAAUGUUAUAUACCCGCUUAGCCCAAGGGUAAUGGGUAUAAAAAGCGCAACAAGAAAAUUCUGUGUGCAGAACACACGCGCUCACAUAUGCAAAUCAAGCAA .----......(((((.((..(((..((((((((.(((.((....)))))))))))))...(((((....(((....)))(((((.....)))))))))).......)))..)).))))) ( -34.00) >DroSim_CAF1 4943 116 - 1 G----CAAAAAUUGCACGAAAUCUAAUAUACCCGCUUAGUCCAAGGGUAAUGGGUAUAAAAAGCGCAACAAGAAAAUUCUGUGUGCAGAACACACACGCUCACAUAUGCAAAUCAAGCAA (----(.....(((((..........((((((((.(((.(.....).)))))))))))...((((.....(((....)))(((((.....))))).))))......))))).....)).. ( -26.50) >DroEre_CAF1 5851 115 - 1 G----CACAAAAUGCUGGAAAUGUGAUAUACCCGCCUUACCCAUGGGUAAGGGGUAUAA-AAGCGCAACAAGAAAAUUCUGUGUGCAGAACACACACGCUCACAUAUGCAAAUCUAGCAA .----.......(((((((..((((.(((((((..((((((....))))))))))))).-..(((((...(((....))).)))))....))))...((........))...))))))). ( -41.20) >DroYak_CAF1 2516 120 - 1 ACAAACAUAAAUUGCUCAAAAUGUAAGAUACCCUCUUUGCCCAAGGGUAAUGGGUAUAAAAAGCGCAACAAGAAAAUUCUGUGUGCAGAACACACACGCUCACAUAUGCAAAUCAAGCAA ...........(((((.....((((..((((((...(((((....))))).))))))....((((.....(((....)))(((((.....))))).))))......)))).....))))) ( -30.40) >consensus G____CAUAAAUUGCUCGAAAUGUAAUAUACCCGCUUAGCCCAAGGGUAAUGGGUAUAAAAAGCGCAACAAGAAAAUUCUGUGUGCAGAACACACACGCUCACAUAUGCAAAUCAAGCAA ............(((...........(((((((.....(((....)))...)))))))....(((((...(((....))).)))))...........((........)).......))). (-22.38 = -22.46 + 0.08)


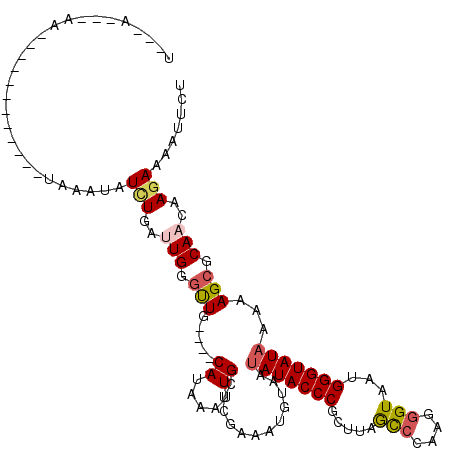
| Location | 13,572,110 – 13,572,226 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.76 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -10.12 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13572110 116 - 22407834 UACUAUUAAACUCUUUUAAAAGUAAAUAUCUAAUUGGGUUG----CAUAAAUUGCACAUAAUUUAAUAUACCCGCUUAGCGCAAGGGUAAUGGGUAUAAAAAGCGCAACAAGAAAAUUCU ((((.(((((....))))).)))).........((..((((----(.(((((((....))))))).((((((((.(((.(.....).)))))))))))......)))))..))....... ( -23.20) >DroSec_CAF1 5439 94 - 1 ----------------------UAAAUAUCUUUUUGGGUUG----CAUAAAUUGCUCGAAAUGUUAUAUACCCGCUUAGCCCAAGGGUAAUGGGUAUAAAAAGCGCAACAAGAAAAUUCU ----------------------......((((.(((.((((----((.....)))...........((((((((.(((.((....)))))))))))))...))).))).))))....... ( -23.50) >DroSim_CAF1 4983 94 - 1 ----------------------UAAAUAUAUGAUUGGGCUG----CAAAAAUUGCACGAAAUCUAAUAUACCCGCUUAGUCCAAGGGUAAUGGGUAUAAAAAGCGCAACAAGAAAAUUCU ----------------------........((.(((.((((----((.....)))...........((((((((.(((.(.....).)))))))))))...))).))))).......... ( -19.90) >DroEre_CAF1 5891 103 - 1 UGACAGC-AAA-----------UAAAGAUUUGGGUGGCUUG----CACAAAAUGCUGGAAAUGUGAUAUACCCGCCUUACCCAUGGGUAAGGGGUAUAA-AAGCGCAACAAGAAAAUUCU ...((((-(..-----------.......((..(((.....----)))..)))))))....((((.(((((((..((((((....))))))))))))).-...))))............. ( -27.20) >DroYak_CAF1 2556 119 - 1 UGACAGC-AAAUCCAUUAAAAAUAAUCAUUUGGAUGCGCUACAAACAUAAAUUGCUCAAAAUGUAAGAUACCCUCUUUGCCCAAGGGUAAUGGGUAUAAAAAGCGCAACAAGAAAAUUCU .......-..(((((...............)))))(((((...........((((.......)))).((((((...(((((....))))).))))))....))))).............. ( -28.36) >consensus U___A___AA____________UAAAUAUCUGAUUGGGUUG____CAUAAAUUGCUCGAAAUGUAAUAUACCCGCUUAGCCCAAGGGUAAUGGGUAUAAAAAGCGCAACAAGAAAAUUCU ............................(((..(((.(((.....((.....))............(((((((.....(((....)))...)))))))...))).)))..)))....... (-10.12 = -10.60 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:21 2006