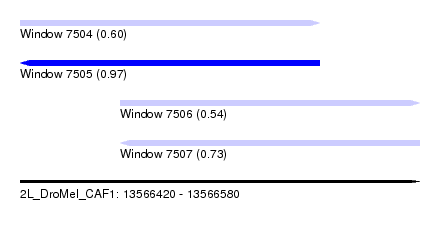
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,566,420 – 13,566,580 |
| Length | 160 |
| Max. P | 0.966050 |

| Location | 13,566,420 – 13,566,540 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -37.80 |
| Energy contribution | -38.67 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13566420 120 + 22407834 AGAGACAGAGCAGACAUUUCUGGAGUUUGAGCUAAAUGCGAAGUGGUGGCAGGAUGGCAGAUCCUGACGAGAAUGCGGGGAUAGCUCCCCAGUUCCCAUGGCCAUCCUGCUAUUUAGCCA .......(.((...((((((...(((....)))......))))))(((((((((((((...((.....))((((..(((((....))))).)))).....)))))))))))))...))). ( -41.80) >DroSec_CAF1 22844 120 + 1 AGAGACAGAGCAGACAUUUCUGGAGUUUGAGCUAAAUGCGAAGUGGUGGCAGGCUGGCAGAUCCUGCCGAGAAUCUGGGGAUAGCUCCCCAGUUCCCAUGGCCAUCCUGCUAUUUAGCCA .....(((((.......))))).....((.((((((((((..(((((((((((.........))))))(.(((.(((((((....)))))))))).)...)))))..))).))))))))) ( -45.10) >DroSim_CAF1 30408 120 + 1 AGAGACAGAGCAGACAUUUCUGGAGUUUGAGCUAAAUGCGAAGUGGUGGCAGGCUGGCAGAUCCUGCCGAGAAUCUGGGGAUAGCUCCCCAGUUCCCAUGGCCAUCCUGCUAUUUAGCCA .....(((((.......))))).....((.((((((((((..(((((((((((.........))))))(.(((.(((((((....)))))))))).)...)))))..))).))))))))) ( -45.10) >DroYak_CAF1 20682 117 + 1 AGAGACAGAGCAGACAUUUCUGGAGUUUGAGCUAAAUGCGAAGUGGUGGCAGGCUGGUAGAUCCUGUCGAGAAU---GGGAUAACUCCCCAGUUCCCAUGGCCAUCCUGCUAUUUAGCCA .....(((((.......))))).....((.((((((((...((.((((((.(((.((.....)).)))....((---((((..(((....))))))))).))))))))..)))))))))) ( -35.50) >consensus AGAGACAGAGCAGACAUUUCUGGAGUUUGAGCUAAAUGCGAAGUGGUGGCAGGCUGGCAGAUCCUGCCGAGAAUCUGGGGAUAGCUCCCCAGUUCCCAUGGCCAUCCUGCUAUUUAGCCA .....(((((.......))))).....((.((((((((((..(((((((((((.........))))))(.((((.((((((....)))))))))).)...)))))..))).))))))))) (-37.80 = -38.67 + 0.88)


| Location | 13,566,420 – 13,566,540 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -38.30 |
| Energy contribution | -39.67 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13566420 120 - 22407834 UGGCUAAAUAGCAGGAUGGCCAUGGGAACUGGGGAGCUAUCCCCGCAUUCUCGUCAGGAUCUGCCAUCCUGCCACCACUUCGCAUUUAGCUCAAACUCCAGAAAUGUCUGCUCUGUCUCU .((((((((.((((((((((.(((((((.((((((....))))))..)))))))........))))))))))...........)))))))).......((((....)))).......... ( -39.50) >DroSec_CAF1 22844 120 - 1 UGGCUAAAUAGCAGGAUGGCCAUGGGAACUGGGGAGCUAUCCCCAGAUUCUCGGCAGGAUCUGCCAGCCUGCCACCACUUCGCAUUUAGCUCAAACUCCAGAAAUGUCUGCUCUGUCUCU .((((((((.(((((.(((....((((((((((((....))))))).)))))((((((.........)))))).)))))).)))))))))).......((((....)))).......... ( -46.50) >DroSim_CAF1 30408 120 - 1 UGGCUAAAUAGCAGGAUGGCCAUGGGAACUGGGGAGCUAUCCCCAGAUUCUCGGCAGGAUCUGCCAGCCUGCCACCACUUCGCAUUUAGCUCAAACUCCAGAAAUGUCUGCUCUGUCUCU .((((((((.(((((.(((....((((((((((((....))))))).)))))((((((.........)))))).)))))).)))))))))).......((((....)))).......... ( -46.50) >DroYak_CAF1 20682 117 - 1 UGGCUAAAUAGCAGGAUGGCCAUGGGAACUGGGGAGUUAUCCC---AUUCUCGACAGGAUCUACCAGCCUGCCACCACUUCGCAUUUAGCUCAAACUCCAGAAAUGUCUGCUCUGUCUCU .((((((((.(((((.(((....(((((...((((....))))---.)))))(.((((.........)))).).)))))).)))))))))).......((((....)))).......... ( -31.20) >consensus UGGCUAAAUAGCAGGAUGGCCAUGGGAACUGGGGAGCUAUCCCCAGAUUCUCGGCAGGAUCUGCCAGCCUGCCACCACUUCGCAUUUAGCUCAAACUCCAGAAAUGUCUGCUCUGUCUCU .((((((((.(((((.(((....((((((((((((....))))))).)))))((((((.........)))))).)))))).)))))))))).......((((....)))).......... (-38.30 = -39.67 + 1.38)


| Location | 13,566,460 – 13,566,580 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -36.67 |
| Energy contribution | -35.90 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13566460 120 + 22407834 AAGUGGUGGCAGGAUGGCAGAUCCUGACGAGAAUGCGGGGAUAGCUCCCCAGUUCCCAUGGCCAUCCUGCUAUUUAGCCAUGUUUUGUUUGUUUUUGUGCUUUGGCUGCAGUUUGAGCAC ..((((((((((((((((...((.....))((((..(((((....))))).)))).....))))))))))).....)))))...............((((((..((....))..)))))) ( -45.60) >DroSim_CAF1 30448 120 + 1 AAGUGGUGGCAGGCUGGCAGAUCCUGCCGAGAAUCUGGGGAUAGCUCCCCAGUUCCCAUGGCCAUCCUGCUAUUUAGCCAUGUUUUGUUUGUUUUUGUGCUUUGGCUGCAGUUUGAGCGC ..(((((((((((..(((((...)))))(.(((.(((((((....)))))))))).)........)))))).....)))))...............((((((..((....))..)))))) ( -43.60) >DroYak_CAF1 20722 117 + 1 AAGUGGUGGCAGGCUGGUAGAUCCUGUCGAGAAU---GGGAUAACUCCCCAGUUCCCAUGGCCAUCCUGCUAUUUAGCCAUGUUUUGUUUGUUUUUGUGCUCUAGCUGCAGUUUGAGUGC ..(((((((((((.((((.(((...)))....((---((((..(((....))))))))).)))).)))))).....)))))...............(..(((.(((....))).)))..) ( -35.30) >consensus AAGUGGUGGCAGGCUGGCAGAUCCUGACGAGAAU__GGGGAUAGCUCCCCAGUUCCCAUGGCCAUCCUGCUAUUUAGCCAUGUUUUGUUUGUUUUUGUGCUUUGGCUGCAGUUUGAGCGC ..(((((((((((.((((...((.....))((((..(((((....))))).)))).....)))).)))))).....)))))...............((((((.(((....))).)))))) (-36.67 = -35.90 + -0.77)



| Location | 13,566,460 – 13,566,580 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -28.51 |
| Energy contribution | -28.18 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13566460 120 - 22407834 GUGCUCAAACUGCAGCCAAAGCACAAAAACAAACAAAACAUGGCUAAAUAGCAGGAUGGCCAUGGGAACUGGGGAGCUAUCCCCGCAUUCUCGUCAGGAUCUGCCAUCCUGCCACCACUU (((..........(((((......................))))).....((((((((((.(((((((.((((((....))))))..)))))))........))))))))))...))).. ( -39.25) >DroSim_CAF1 30448 120 - 1 GCGCUCAAACUGCAGCCAAAGCACAAAAACAAACAAAACAUGGCUAAAUAGCAGGAUGGCCAUGGGAACUGGGGAGCUAUCCCCAGAUUCUCGGCAGGAUCUGCCAGCCUGCCACCACUU (.((.((..(((((((((......................))))).....))))..)))))..((((((((((((....))))))).)))))((((((.........))))))....... ( -43.15) >DroYak_CAF1 20722 117 - 1 GCACUCAAACUGCAGCUAGAGCACAAAAACAAACAAAACAUGGCUAAAUAGCAGGAUGGCCAUGGGAACUGGGGAGUUAUCCC---AUUCUCGACAGGAUCUACCAGCCUGCCACCACUU (((.......))).((((.(((.((...............)))))...)))).((.((((..((((((...((((....))))---.))))))..(((.........))))))))).... ( -29.26) >consensus GCGCUCAAACUGCAGCCAAAGCACAAAAACAAACAAAACAUGGCUAAAUAGCAGGAUGGCCAUGGGAACUGGGGAGCUAUCCCC__AUUCUCGACAGGAUCUGCCAGCCUGCCACCACUU (((.......)))(((((......................))))).....(((((.(((((.((((((..(((((....)))))...))))))...)).....))).)))))........ (-28.51 = -28.18 + -0.33)

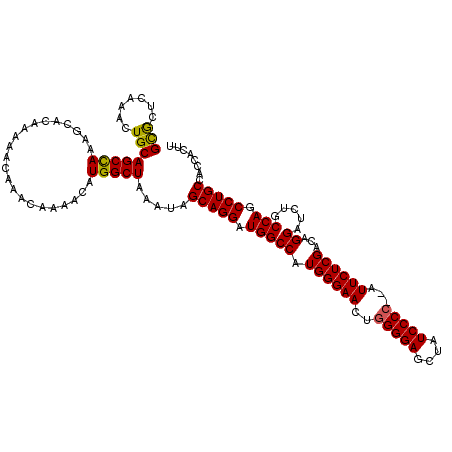
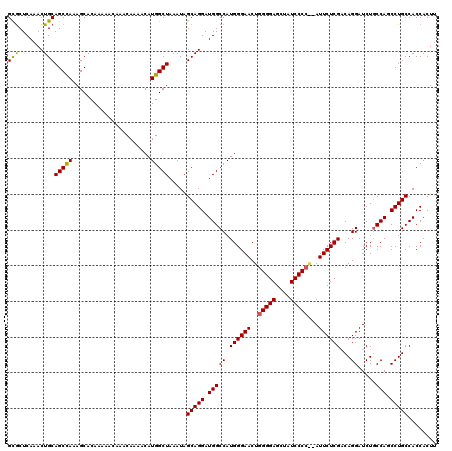
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:09 2006