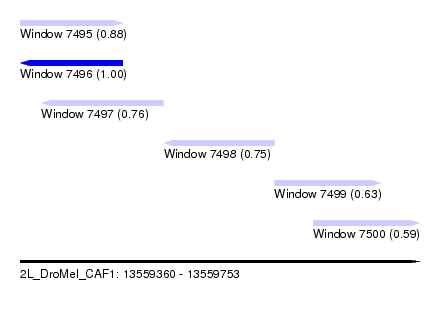
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,559,360 – 13,559,753 |
| Length | 393 |
| Max. P | 0.995771 |

| Location | 13,559,360 – 13,559,461 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -26.68 |
| Energy contribution | -26.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13559360 101 + 22407834 G-AGUAGUUGGAAAAAUGCCCGACGUCGAGUAGCUCUAGAAUUCGUUUAGGCUGUCCCCACUCUUGCUAGCCAGCCCACUCGAAACACCGGGCUUUUCCUCA .-.......((((((..(((((...(((((((((.(((((.....))))))))............(((....)))..)))))).....)))))))))))... ( -29.60) >DroSec_CAF1 20560 101 + 1 G-AGUAGCUGGAAAAAUGCCCGACGUCGAGUAGCUCUAGAAUUCGUUUAGGCUGUCCCCACUCUUGCUAGCCAGCCCACUCGAAAGCCCGGGCUUUUCCUCA .-.......((((((..(((((...(((((((((.(((((.....))))))))............(((....)))..)))))).....)))))))))))... ( -29.60) >DroSim_CAF1 28126 101 + 1 G-AGUAGCUGGAAAAAUGCCCGACGUCGAGUAGCUCUAGAAUUCGUUUAGGCUGUCCCCACUCUUGCUAGCCAGCCCACUCGAAACCCCGGGCUUUUCCUCA .-.......((((((..(((((...(((((((((.(((((.....))))))))............(((....)))..)))))).....)))))))))))... ( -29.60) >DroEre_CAF1 19940 101 + 1 A-AGUAGUUGGAAAAAUGCCCGACGUCGAGUAGCUCUAGAAUUCGUUUAGGCUGGCCCCACUCUUGCGAGCCAACCCACUCGAAACCCCGGGCUUUUCCUCA .-.......((((((..(((((...(((((((((.(((((.....))))))))(((.(((....)).).))).....)))))).....)))))))))))... ( -32.90) >DroYak_CAF1 18341 101 + 1 AGAGUAGUUGGAAAAAUGCCCGACGUCGAGUAGCUCUAGAAUUCGUUUAGGCUGGC-CCACUCUUACGAGCCAACCCACUCGAAACUCCGGGCUUUUCCUCA .........((((((..(((((...(((((((((.(((((.....))))))))(((-.(........).))).....)))))).....)))))))))))... ( -32.60) >consensus G_AGUAGUUGGAAAAAUGCCCGACGUCGAGUAGCUCUAGAAUUCGUUUAGGCUGUCCCCACUCUUGCUAGCCAGCCCACUCGAAACCCCGGGCUUUUCCUCA .........((((((..(((((...(((((((((.(((((.....)))))))).......((......)).......)))))).....)))))))))))... (-26.68 = -26.68 + -0.00)

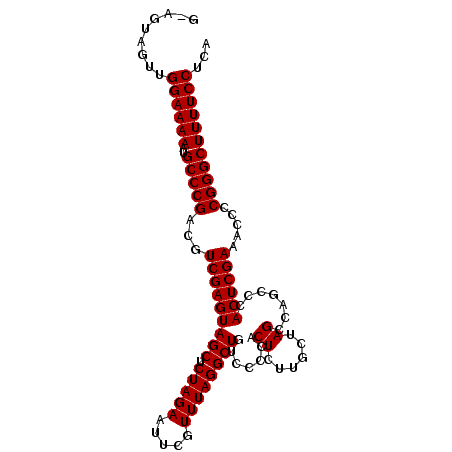
| Location | 13,559,360 – 13,559,461 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -39.72 |
| Consensus MFE | -32.80 |
| Energy contribution | -32.96 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13559360 101 - 22407834 UGAGGAAAAGCCCGGUGUUUCGAGUGGGCUGGCUAGCAAGAGUGGGGACAGCCUAAACGAAUUCUAGAGCUACUCGACGUCGGGCAUUUUUCCAACUACU-C ...(((((((((((..((((((..(((((((.((.((....))..)).)))))))..)))......(((...))))))..)))))..)))))).......-. ( -37.90) >DroSec_CAF1 20560 101 - 1 UGAGGAAAAGCCCGGGCUUUCGAGUGGGCUGGCUAGCAAGAGUGGGGACAGCCUAAACGAAUUCUAGAGCUACUCGACGUCGGGCAUUUUUCCAGCUACU-C ...(((((((((((((..((((..(((((((.((.((....))..)).)))))))..))))..)).(((...))).....)))))..)))))).......-. ( -35.80) >DroSim_CAF1 28126 101 - 1 UGAGGAAAAGCCCGGGGUUUCGAGUGGGCUGGCUAGCAAGAGUGGGGACAGCCUAAACGAAUUCUAGAGCUACUCGACGUCGGGCAUUUUUCCAGCUACU-C ...((((((((((((.((((((..(((((((.((.((....))..)).)))))))..)))......(((...)))))).))))))..)))))).......-. ( -38.00) >DroEre_CAF1 19940 101 - 1 UGAGGAAAAGCCCGGGGUUUCGAGUGGGUUGGCUCGCAAGAGUGGGGCCAGCCUAAACGAAUUCUAGAGCUACUCGACGUCGGGCAUUUUUCCAACUACU-U ...((((((((((((.((((((..(((((((((((.((....)))))))))))))..)))......(((...)))))).))))))..)))))).......-. ( -44.00) >DroYak_CAF1 18341 101 - 1 UGAGGAAAAGCCCGGAGUUUCGAGUGGGUUGGCUCGUAAGAGUGG-GCCAGCCUAAACGAAUUCUAGAGCUACUCGACGUCGGGCAUUUUUCCAACUACUCU ...(((((((((((..((((((..((((((((((((......)))-)))))))))..)))......(((...))))))..)))))..))))))......... ( -42.90) >consensus UGAGGAAAAGCCCGGGGUUUCGAGUGGGCUGGCUAGCAAGAGUGGGGACAGCCUAAACGAAUUCUAGAGCUACUCGACGUCGGGCAUUUUUCCAACUACU_C ...((((((((((((.(..((((((((.((((.......(..((((.....))))..).....))))..))))))))).))))))..))))))......... (-32.80 = -32.96 + 0.16)


| Location | 13,559,381 – 13,559,501 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.04 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13559381 120 - 22407834 UGACAAAGGUGGAAAAAUGGUUGGAAAAACGUGGACGAGUUGAGGAAAAGCCCGGUGUUUCGAGUGGGCUGGCUAGCAAGAGUGGGGACAGCCUAAACGAAUUCUAGAGCUACUCGACGU ...(((..((......))..)))......((....)).((((((....((((..(...((((..(((((((.((.((....))..)).)))))))..))))..)..).))).)))))).. ( -31.90) >DroSec_CAF1 20581 120 - 1 UGACAAAGGUGGAAAACUGGUUGGAAAAACGUGAACGAGUUGAGGAAAAGCCCGGGCUUUCGAGUGGGCUGGCUAGCAAGAGUGGGGACAGCCUAAACGAAUUCUAGAGCUACUCGACGU ...(((.(((.....)))..)))......((....)).((((((....(((.(.((..((((..(((((((.((.((....))..)).)))))))..))))..)).).))).)))))).. ( -35.60) >DroSim_CAF1 28147 120 - 1 CGACAAAGGUGAAAAACUGGUUGGAAAAACGCGGACGAGUUGAGGAAAAGCCCGGGGUUUCGAGUGGGCUGGCUAGCAAGAGUGGGGACAGCCUAAACGAAUUCUAGAGCUACUCGACGU ((((...(((.....))).))))...............((((((....((((..(((((.((..(((((((.((.((....))..)).)))))))..)))))))..).))).)))))).. ( -36.60) >DroEre_CAF1 19961 118 - 1 UGACAAAGGUGGA-AAAUGGUUGGAA-AACGUGGACGAGUUGAGGAAAAGCCCGGGGUUUCGAGUGGGUUGGCUCGCAAGAGUGGGGCCAGCCUAAACGAAUUCUAGAGCUACUCGACGU .............-.....(((....-)))........((((((....((((..(((((.((..(((((((((((.((....)))))))))))))..)))))))..).))).)))))).. ( -39.60) >DroYak_CAF1 18363 118 - 1 UGACAAAGGUGGAGAAAUGGUUGGAA-AACGUGAACGAGUUGAGGAAAAGCCCGGAGUUUCGAGUGGGUUGGCUCGUAAGAGUGG-GCCAGCCUAAACGAAUUCUAGAGCUACUCGACGU ...(((..((......))..)))...-..((....)).((((((....((((..(((((.((..((((((((((((......)))-)))))))))..)))))))..).))).)))))).. ( -41.70) >consensus UGACAAAGGUGGAAAAAUGGUUGGAAAAACGUGGACGAGUUGAGGAAAAGCCCGGGGUUUCGAGUGGGCUGGCUAGCAAGAGUGGGGACAGCCUAAACGAAUUCUAGAGCUACUCGACGU .............................((....)).((((((....((((..(((((.((..((((((((((......))).....)))))))..)))))))..).))).)))))).. (-28.00 = -28.04 + 0.04)

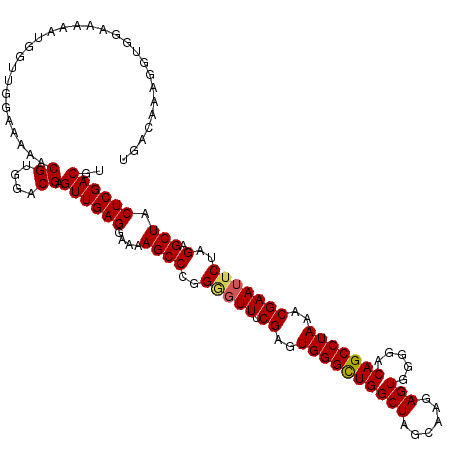
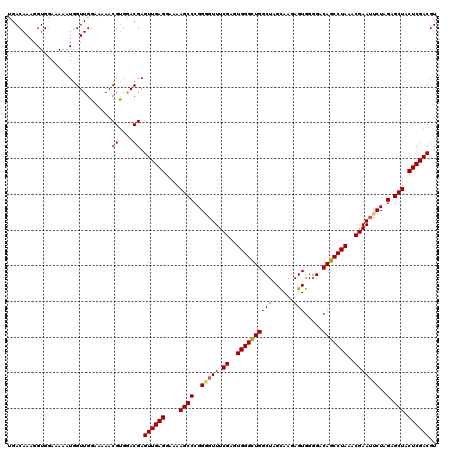
| Location | 13,559,501 – 13,559,610 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.28 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -20.94 |
| Energy contribution | -22.86 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13559501 109 - 22407834 UUUUGCAGUUCUUUGCGAUUUUUCCUUUUUGCCCGUCAGUCAACUGACAAGUCAACAGCGAUGCAACCUGCAAAAGGCAAGGGCUGAGGAGGAGAAAAAAAAAAGAUGG ..((((((....))))))(((((((((((((((((((((....))))).........((..(((.....)))....))..)))).))))))))))))............ ( -35.70) >DroSec_CAF1 20701 106 - 1 UUUUGCAGUUCUUUGCGAUUUUUCGUUUUGGCCCGUCAGUCAACUGACAAGUCAACAGCGAUGCAACCUGCAAAAGGCAAGGGCUGAGGAGGAAAAAAAAA---AAUUG ..((((((....))))))((((((.(((..(((((((((....))))).........((..(((.....)))....))..))))..))).)))))).....---..... ( -31.90) >DroSim_CAF1 28267 108 - 1 UUUUGCAGUUCUUUGCGAUUUUUCCUUUUUGCCCGUCAGUCAACUGACAAGUCAACAGCAAUGCAACCUGCAAAAGGCAAGGGCUGAGGAGGAAAAAAAAA-AGAAUGG .......(((((((....(((((((((((((((((((((....))))).........((..(((.....)))....))..)))).))))))))))))..))-))))).. ( -37.30) >DroEre_CAF1 20079 104 - 1 U-UUACAGUUCCUUGC-AUUUUUCCUUUUUGGCCGUCACUCAACUGACAAGUCAACAGCGAUGCAACCUGCAAAAGGCAAGGGCUGAGGAGAGAAAAAGAA---AAAGG .-........((((..-.(((((((((((..(((((((......)))).........((..(((.....)))....))...)))..))))).))))))...---.)))) ( -28.60) >DroYak_CAF1 18481 97 - 1 UUUUACAGCUCUUUGC-AUUUUUUCUUUUUGCCCGUCACUCAACUGACAAGUCAACAGCGAUGCAACCUGCAAAAGGGAAG-GCUGAGGAGC-------AA---AGAGG .......(((((((((-....(((((((((((..((((......))))..(((......))).......))))))))))).-)).)))))))-------..---..... ( -26.60) >consensus UUUUGCAGUUCUUUGCGAUUUUUCCUUUUUGCCCGUCAGUCAACUGACAAGUCAACAGCGAUGCAACCUGCAAAAGGCAAGGGCUGAGGAGGAAAAAAAAA___AAUGG ..((((((....))))))(((((((((...(((((((((....)))))..(((....(((........)))....)))..)))).)))))))))............... (-20.94 = -22.86 + 1.92)

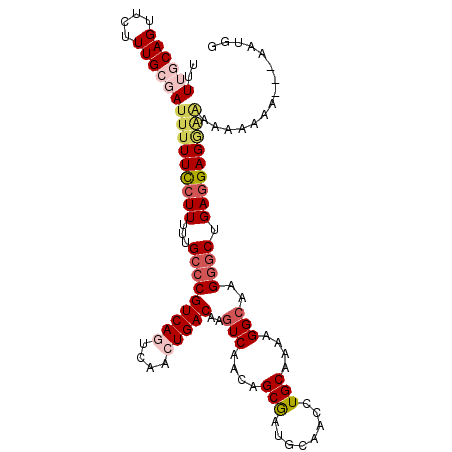
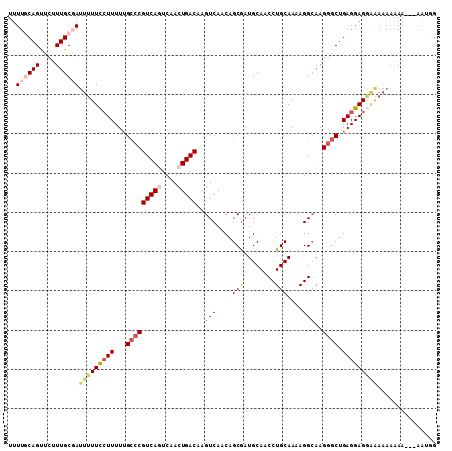
| Location | 13,559,610 – 13,559,715 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 88.07 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -17.31 |
| Energy contribution | -18.30 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13559610 105 + 22407834 GUAGUGCCUUUUAUUCUGUUUUGUUUUUUUU--UCCCCCGCUCAUGCAAAUGAUUGGGUCACUCGACAAGGCGUGGCACACAUGUAUUAAAUAUAUGUAU-GUAUGUA ...(((((........(((((((((......--...((((.((((....)))).))))......))))))))).)))))((((((((...))))))))..-....... ( -26.06) >DroSec_CAF1 20807 107 + 1 GUAGUGGCUUUUAUUCGGUUUUGUUUUUUUUUUUUCCCCGCACAUGCAAAUGUGUGGGUCACUCGACAAGGCGUGGCAUACAUGUUUUAAAUAUAUGUAU-GUAUGUA (.((((((........((.................))((((((((....)))))))))))))))......(((((.(((((((((.......))))))))-)))))). ( -31.03) >DroSim_CAF1 28375 103 + 1 GUAGUGCCUUUUAUUCUGUUUUGUUUUUUUU----UCCCGCACAUGCAAAUGUUUGGGUCACUCGACAAGGCGUGGCAUACAUGUAUUAAAUAUAUGUAU-GUAUGUA .................((((((((......----.((((.((((....)))).))))......))))))))...((((((((((((...))))))))))-))..... ( -27.62) >DroEre_CAF1 20183 100 + 1 GUAGUGCCUUUUAUUCUGUUUUGUUUUU--------GCCGCUCAUGCAAAUGUUUGGGUCACUCGACAAGGCGUGGCACACAUGUAUUAAAUAUAUAUCGAACAUAUA ((.(((((........(((((((((...--------(..(((((.((....)).)))))..)..))))))))).)))))))((((((.....)))))).......... ( -21.30) >consensus GUAGUGCCUUUUAUUCUGUUUUGUUUUUUUU____CCCCGCACAUGCAAAUGUUUGGGUCACUCGACAAGGCGUGGCACACAUGUAUUAAAUAUAUGUAU_GUAUGUA ...(((((........(((((((((...........((((.((((....)))).))))......))))))))).)))))(((((((.....))))))).......... (-17.31 = -18.30 + 1.00)

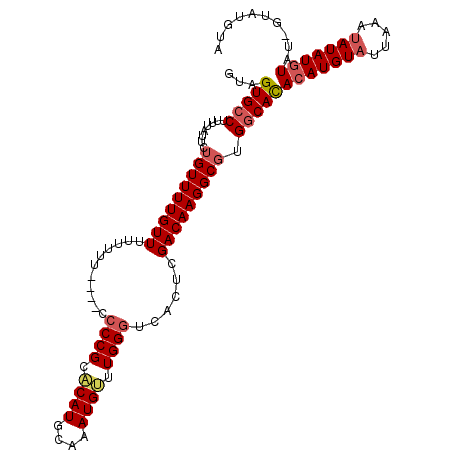
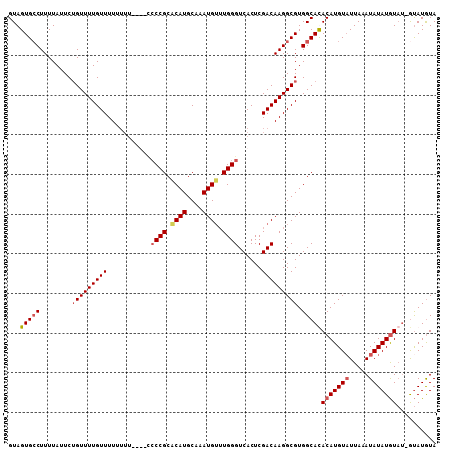
| Location | 13,559,648 – 13,559,753 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -9.42 |
| Energy contribution | -9.92 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13559648 105 + 22407834 CUCAUGCAAAUGAU--UGGGUCACUCGACAAGGCGUGGCACACAUGUAUUAAAUAUAUGU-AU-GUAUGUA----------UAUAAAAUUGUUCGA-UUUUUUAUGAUUCAAUUUUAUGC .....(((...(((--((((((..........(((((.((.((((((((...))))))))-.)-))))))(----------((.((((((....))-)))).))))))))))))...))) ( -23.10) >DroPse_CAF1 50211 120 + 1 UUCAUGGAAAUGUCGGAGGGACACACGUAGAGGCGUGGCACAUAUGCAUGUGGCACAUACUCGCACUCGUAAUCGUUGUCUUAUUUUAAUAUUCGACUUUUUUAUGAUUCAAUUUUAUGC .(((((((((.((((((((((((.(((..((((((((.(((((....))))).)))......)).))).....))))))))).........)))))).)))))))))............. ( -34.10) >DroSec_CAF1 20847 105 + 1 CACAUGCAAAUGUG--UGGGUCACUCGACAAGGCGUGGCAUACAUGUUUUAAAUAUAUGU-AU-GUAUGUA----------UAUAAAAUUAUUCGA-UUUUUUAUGAUUCAAUUUUAUGC .(((((((.(((((--((.(((....)))((((((((.....))))))))...)))))))-.)-)))))).----------(((((((((..(((.-.......)))...))))))))). ( -24.00) >DroSim_CAF1 28411 105 + 1 CACAUGCAAAUGUU--UGGGUCACUCGACAAGGCGUGGCAUACAUGUAUUAAAUAUAUGU-AU-GUAUGUA----------UAUAAAGUUGUUCGA-UUUUUUAUGAUUCAAUUUUAUGC ..(((((...((((--.((....)).))))..)))))((((((((((((...))))))))-))-)).....----------((((((((((.(((.-.......)))..)))))))))). ( -29.10) >DroEre_CAF1 20215 106 + 1 CUCAUGCAAAUGUU--UGGGUCACUCGACAAGGCGUGGCACACAUGUAUUAAAUAUAUAU-CGAACAUAUA----------UAUGUUGCUGUUCGA-UUUUUUAUGAUUCAAUUUUAUGC .....(((.....(--(((((((.........(((((.....)))))...........((-((((((((..----------.....)).)))))))-)......)))))))).....))) ( -20.80) >consensus CUCAUGCAAAUGUU__UGGGUCACUCGACAAGGCGUGGCACACAUGUAUUAAAUAUAUGU_AU_GUAUGUA__________UAUAAAAUUGUUCGA_UUUUUUAUGAUUCAAUUUUAUGC .((((....))))....((((((.........(((((.....))))).....((((((......))))))..................................)))))).......... ( -9.42 = -9.92 + 0.50)

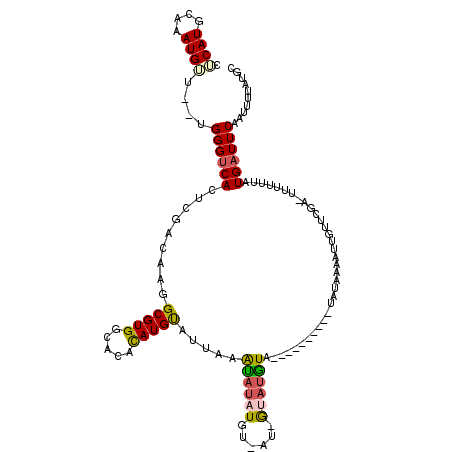
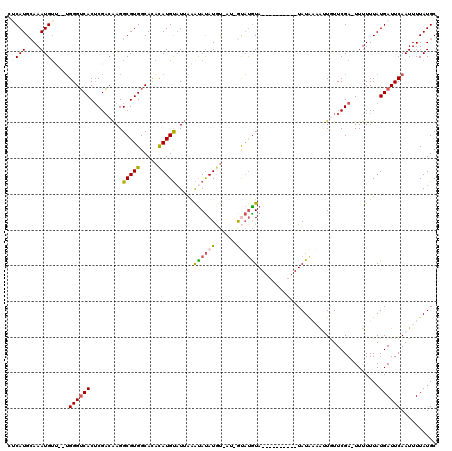
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:02 2006