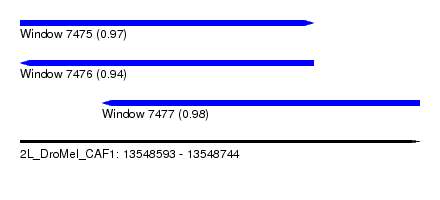
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,548,593 – 13,548,744 |
| Length | 151 |
| Max. P | 0.978104 |

| Location | 13,548,593 – 13,548,704 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 96.23 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -37.72 |
| Energy contribution | -37.48 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13548593 111 + 22407834 GUGGCAGAGCCGAGGUGCCGUGGAAUGGGGA-UUUUCCCGAAAAGUAACGGCUCUUUCGGGAAAAACCGUUUUCCGUAUUGAUUUCGGGGGGCGCUGCAUGUGAGCUUCCCA ..(((...)))(((((....(((((.(.((.-((((((((((((((....))).))))))))))).)).).))))).....)))))(((((((.(.......).))))))). ( -42.10) >DroSec_CAF1 7453 111 + 1 GUGGCAGAGCCGAGGAGCCGUGGAAUGGGGA-UUUUCCCGAACAGUAACGGCUCUUUCGGGAAAAACCGUUUUCCGUAUUGAUUUCGGGGGGCGCUGCAUGUGAGCUUCCCA ..(((...)))((((((((((.(..(((((.-...)))))..)....)))))))))).(((((.....(((..(((.........)))..)))(((.......)))))))). ( -41.50) >DroSim_CAF1 7679 112 + 1 GUGGCAGAGCCGAGGAGCCGUGGAAUGGGUACUUUUCCCGAAAAGUAACGGCUCUUUCGGGAAAAACCGUUUUCCGUAUUGAUUUCGGGGGGCGCUGCAUGUGAGCUUCCCA ..(((...)))((((((((((....((((.......)))).......)))))))))).(((((.....(((..(((.........)))..)))(((.......)))))))). ( -40.40) >DroEre_CAF1 9795 110 + 1 GUGGCAGAGCCGAGGAGCAAUGGAACGGGGA-UU-UCCCGAAAAGUAACGGCUCUUUCGGGAAAAACCGUUUUCCGUAUUGAUUUCGGGGGGCGCUGCAUGUGAGCUUCCCA .((((...)))).(((((((((((((((...-((-(((((((((((....))).))))))))))..))))..)))))..)).))))(((((((.(.......).))))))). ( -42.30) >DroYak_CAF1 7771 111 + 1 GUGGCAGAGCCGAGGAGCAAUGGAACGGGGA-UUUUCCCGAAAAGUAACGGCUCUUUCGGGAAAAACCGUUUUCCGUAUUGAUUUCGGGGGGCGCUGCAUGUGAGCUUUCCA .(((.(((((((.(.(((..........((.-((((((((((((((....))).))))))))))).))(((..(((.........)))..)))))).)...)).)))))))) ( -41.80) >consensus GUGGCAGAGCCGAGGAGCCGUGGAAUGGGGA_UUUUCCCGAAAAGUAACGGCUCUUUCGGGAAAAACCGUUUUCCGUAUUGAUUUCGGGGGGCGCUGCAUGUGAGCUUCCCA ..(((...)))((((....(((((((((....((((((((((((((....))).))))))))))).))))..))))).....))))(((((((.(.......).))))))). (-37.72 = -37.48 + -0.24)

| Location | 13,548,593 – 13,548,704 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 96.23 |
| Mean single sequence MFE | -33.09 |
| Consensus MFE | -28.76 |
| Energy contribution | -29.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13548593 111 - 22407834 UGGGAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUUUUCGGGAAAA-UCCCCAUUCCACGGCACCUCGGCUCUGCCAC .(((..((((....).)))..))).............((((...((.((((((((((.(((......)))))))))))))-.))...))))..((((.........)))).. ( -33.10) >DroSec_CAF1 7453 111 - 1 UGGGAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUGUUCGGGAAAA-UCCCCAUUCCACGGCUCCUCGGCUCUGCCAC .(((..((((....).)))..)))(((.........))).....(((.....((((..(((((((........(((....-.)))......))))))).))))....))).. ( -33.04) >DroSim_CAF1 7679 112 - 1 UGGGAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUUUUCGGGAAAAGUACCCAUUCCACGGCUCCUCGGCUCUGCCAC .(((..((((....).)))..)))(((.........))).....(((.....((((..((((((((((((((....)))))))........))))))).))))....))).. ( -32.70) >DroEre_CAF1 9795 110 - 1 UGGGAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUUUUCGGGA-AA-UCCCCGUUCCAUUGCUCCUCGGCUCUGCCAC .(((..((((....).)))..)))((((...((((..((((...((..(((((((((.(((......))))))))))-))-..))..))))))))....))))......... ( -33.80) >DroYak_CAF1 7771 111 - 1 UGGAAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUUUUCGGGAAAA-UCCCCGUUCCAUUGCUCCUCGGCUCUGCCAC ..............((((.(((....((...((((..((((...((.((((((((((.(((......)))))))))))))-.))...)))))))).))...))).))))... ( -32.80) >consensus UGGGAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUUUUCGGGAAAA_UCCCCAUUCCACGGCUCCUCGGCUCUGCCAC ..............((((.(((....(....).....((((...((.((((((((((.(((......)))))))))))))...))..))))..........))).))))... (-28.76 = -29.16 + 0.40)

| Location | 13,548,624 – 13,548,744 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -33.99 |
| Energy contribution | -34.39 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13548624 120 - 22407834 ACUCCUUUCAUCGGGCGUUUUAGAAUGCAUAUUUGGUUUAUGGGAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUUUUCGGGAAAA ............((((((.......(((((....(((((.....)))))...)))))))))))(((((((((......)).((((((((((.....))))))))))...))))))).... ( -35.21) >DroSec_CAF1 7484 120 - 1 ACUCCUUUCAUCGGGCGUUUUAGAAUGCAUAUUUGGUUUAUGGGAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUGUUCGGGAAAA ............((((((.......(((((....(((((.....)))))...)))))))))))(((((.......((((..((((((((((.....)))))))))).))))))))).... ( -35.12) >DroSim_CAF1 7711 120 - 1 ACUCCUUUCAUCGGGCGUUUUAGAAUGCAUAUUUGGUUUAUGGGAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUUUUCGGGAAAA ............((((((.......(((((....(((((.....)))))...)))))))))))(((((((((......)).((((((((((.....))))))))))...))))))).... ( -35.21) >DroEre_CAF1 9826 119 - 1 ACUCCUUUCAUCGGGCGUUUUAGAAUGCAUAUUUCGUUUAUGGGAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUUUUCGGGA-AA ............(((((((......(.((((.......)))).)..((......)))))))))(((((((((......)).((((((((((.....))))))))))...))))))).-.. ( -34.20) >DroYak_CAF1 7802 120 - 1 ACUCCUUUCAUCGGGCGUUUUAGAAUGCAUAUUUGGUUUAUGGAAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUUUUCGGGAAAA ............((((((.......(((((....(((((.....)))))...)))))))))))(((((((((......)).((((((((((.....))))))))))...))))))).... ( -35.11) >consensus ACUCCUUUCAUCGGGCGUUUUAGAAUGCAUAUUUGGUUUAUGGGAAGCUCACAUGCAGCGCCCCCCGAAAUCAAUACGGAAAACGGUUUUUCCCGAAAGAGCCGUUACUUUUCGGGAAAA ............((((((.......(((((....(((((.....)))))...)))))))))))(((((((((......)).((((((((((.....))))))))))...))))))).... (-33.99 = -34.39 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:40 2006