
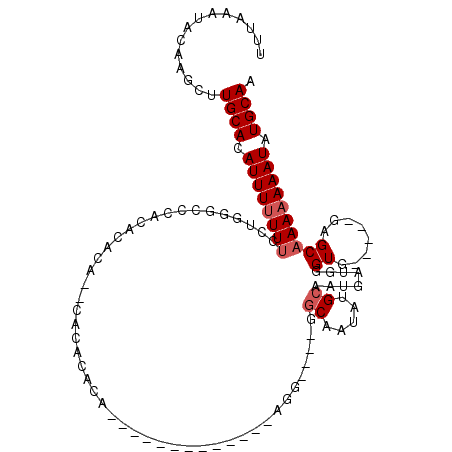
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,534,619 – 13,534,714 |
| Length | 95 |
| Max. P | 0.999870 |

| Location | 13,534,619 – 13,534,714 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.74 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -6.70 |
| Energy contribution | -7.30 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.28 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13534619 95 + 22407834 UUUAAAUACAAGCUUGCACAUUUUUUUCCUGGGCCCACACUCACACUCACACA-------------AGG----GGCAAUAUGCAAUGAUGGGUG-----GAGCAAAAAAAUAUGCAA ..............((((.((((((((.((..(((((((......(((.....-------------.))----)((.....))..)).))))).-----.)).)))))))).)))). ( -22.60) >DroSec_CAF1 20645 93 + 1 UUUAAAUACAAGCUUGCACAUUUUUUUCCUGGGCCCACACACU--CACACACA-------------AGG----GGCAAUAUGCAAUGAUGGGUG-----GAGCAAAAAAAUAUGCAA ..............((((.((((((((.((..(((((((..((--(.......-------------.))----)((.....))..)).))))).-----.)).)))))))).)))). ( -22.40) >DroSim_CAF1 17368 93 + 1 UUUAAAUACAAGCUUGCACAUUUUUUUCCUGAGCCCACACACC--CACACAGA-------------AGG----GGCAAUAUGCAAUGAUGGGUG-----GAGCAAAAAAAUAUGCAA ..............((((.((((((((.((..(((((((..((--(.......-------------.))----)((.....))..)).))))).-----.)).)))))))).)))). ( -25.80) >DroEre_CAF1 20807 110 + 1 UUUAAAUACAAACUUGCACAUUU-UUUCCUGGGCUCACACUCACGCCCACACACACACACAAACACAGGAGCAGGCAAUAUGCAAUGAUGGGUG------AGCAAAAAAAUAUGCAA ..............((((.((((-(((.....((((((..(((.((((...................)).))..((.....))..)))...)))------))).))))))).)))). ( -22.71) >DroAna_CAF1 22989 83 + 1 UUUAAAUACACAUAUGCAUAUUUUUUCACCGUCUCCCCA----------------------------------GCCAAUAUGCCAAGGAGAGUGGGAGGGCGCAAAAAAAUAUGCAA ..............((((((((((((...((((..((((----------------------------------.((..........))....))))..))))..)))))))))))). ( -25.30) >consensus UUUAAAUACAAGCUUGCACAUUUUUUUCCUGGGCCCACACACA__CACACACA_____________AGG____GGCAAUAUGCAAUGAUGGGUG_____GAGCAAAAAAAUAUGCAA ..............((((.((((((((...............................................((.....))........((........)))))))))).)))). ( -6.70 = -7.30 + 0.60)


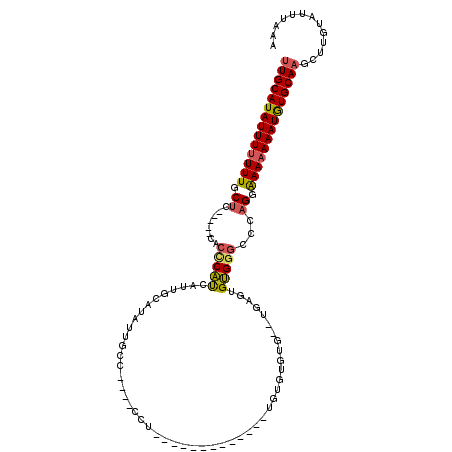
| Location | 13,534,619 – 13,534,714 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.74 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -13.33 |
| Energy contribution | -13.41 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.44 |
| SVM decision value | 4.32 |
| SVM RNA-class probability | 0.999870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13534619 95 - 22407834 UUGCAUAUUUUUUUGCUC-----CACCCAUCAUUGCAUAUUGCC----CCU-------------UGUGUGAGUGUGAGUGUGGGCCCAGGAAAAAAAUGUGCAAGCUUGUAUUUAAA ((((((((((((((.((.-----..(((((..(..(((.(..(.----...-------------...)..))))..)..)))))...)).))))))))))))))............. ( -29.00) >DroSec_CAF1 20645 93 - 1 UUGCAUAUUUUUUUGCUC-----CACCCAUCAUUGCAUAUUGCC----CCU-------------UGUGUGUG--AGUGUGUGGGCCCAGGAAAAAAAUGUGCAAGCUUGUAUUUAAA ((((((((((((((.((.-----..(((((((((((((((....----...-------------.)))))).--)))).)))))...)).))))))))))))))............. ( -30.80) >DroSim_CAF1 17368 93 - 1 UUGCAUAUUUUUUUGCUC-----CACCCAUCAUUGCAUAUUGCC----CCU-------------UCUGUGUG--GGUGUGUGGGCUCAGGAAAAAAAUGUGCAAGCUUGUAUUUAAA ((((((((((((((.((.-----..((((.(((........(((----((.-------------.....).)--))))))))))...)).))))))))))))))............. ( -28.20) >DroEre_CAF1 20807 110 - 1 UUGCAUAUUUUUUUGCU------CACCCAUCAUUGCAUAUUGCCUGCUCCUGUGUUUGUGUGUGUGUGUGGGCGUGAGUGUGAGCCCAGGAAA-AAAUGUGCAAGUUUGUAUUUAAA (((((((((((((((((------((((((.(((.((((((.((..((......))..))))))))))))))((....))))))))....))))-))))))))))............. ( -34.60) >DroAna_CAF1 22989 83 - 1 UUGCAUAUUUUUUUGCGCCCUCCCACUCUCCUUGGCAUAUUGGC----------------------------------UGGGGAGACGGUGAAAAAAUAUGCAUAUGUGUAUUUAAA .((((((((((((..((......)..((((((..((......))----------------------------------..))))))..)..)))))))))))).............. ( -29.50) >consensus UUGCAUAUUUUUUUGCUC_____CACCCAUCAUUGCAUAUUGCC____CCU_____________UGUGUGUG__UGAGUGUGGGCCCAGGAAAAAAAUGUGCAAGCUUGUAUUUAAA ((((((((((((((.((........(((((.................................................)))))...)).))))))))))))))............. (-13.33 = -13.41 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:29 2006