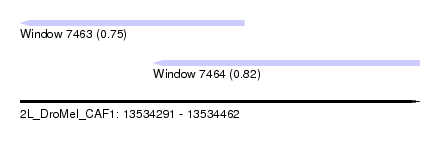
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,534,291 – 13,534,462 |
| Length | 171 |
| Max. P | 0.817806 |

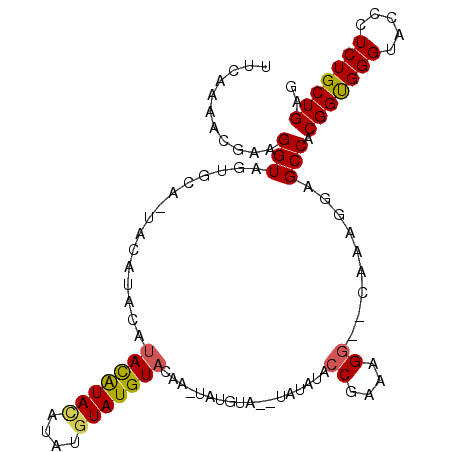
| Location | 13,534,291 – 13,534,387 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -14.87 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13534291 96 - 22407834 UUCAAAACGAAGGUAGUGCAAUACAUUCAUACAUACAUAUGUAUGUACAA---------UAUAUACCGAAAGG--CAAAGGAGCCACGGCGGGUACCCUCUGCUGAG (((.....(((.(((......))).))).(((((((....)))))))...---------........))).((--(......))).(((((((.....))))))).. ( -24.00) >DroSim_CAF1 17056 95 - 1 UUCAAAACGAAGGUAGUGCA-----UACAAACAUACA----UAUGUACAA-UAUGUAUGUAUAUACCGAAAGG--CAAAGGAGCCACGGUGGGUCCCCUCUGCUGAG ...........((((((((.-----.....(((((((----(((.....)-)))))))))......(....))--))..(((.((.....)).)))...)))))... ( -26.60) >DroEre_CAF1 20494 105 - 1 UACAAAACGAAGGUACUGCA-UACAUACAUAUGUAUGUAUGUAUGUACGAGUAUGUAC-AAUAUAACGAAAGGAGCAAAGGAGCCACGGUGGGUGCCCUCUGCUGAG ((((..((....((((((((-(((((((....))))))))))).))))..)).)))).-.......(....).((((.(((.(((......)))..))).))))... ( -32.80) >consensus UUCAAAACGAAGGUAGUGCA_UACAUACAUACAUACAUAUGUAUGUACAA_UAUGUA__UAUAUACCGAAAGG__CAAAGGAGCCACGGUGGGUACCCUCUGCUGAG ...........(((...............(((((((....)))))))..................((....)).........))).(((((((.....))))))).. (-14.87 = -15.10 + 0.23)


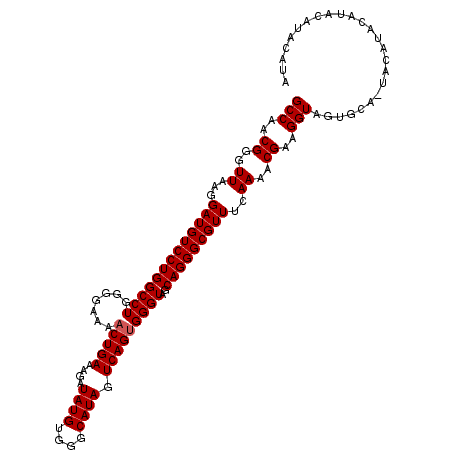
| Location | 13,534,348 – 13,534,462 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.78 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -27.68 |
| Energy contribution | -27.93 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13534348 114 - 22407834 GCCAACGGGUUAAGGAUGUCCUGGCCUGGGGAAAACUGAAAGAUAUGUGGGCAUAGUCAGUGGGUAGCAGGGCGUUUCAAAACGAAGGUAGUGCAAUACAUUCAUACAUACAUA (((..(((((((.((....)))))))))......(((((....((((....)))).))))).))).(((..((.((((.....))))))..))).................... ( -28.70) >DroSim_CAF1 17119 107 - 1 GCCAACGGGUUAAGGAUGUCCUGGCCUGGGGAAAACUGAAAGAUAUGUGGGCAUAGUCAGUGGGUAGCAGGGCGUUUCAAAACGAAGGUAGUGCA-----UACAAACAUACA-- .((..(((((((.((....)))))))))......(((((....((((....)))).)))))))((((((..((.((((.....))))))..))).-----))).........-- ( -29.10) >DroEre_CAF1 20561 113 - 1 GCCAACGGGUUAAGGAUGUCCUGGCCUGGGGAAACCUGAAAGAUAUGUGGGCAUAGUCAGUGGGUAGCAGGGCGUUACAAAACGAAGGUACUGCA-UACAUACAUAUGUAUGUA (((..((..((...((((((((((((((((....)))....((((((....))).)))...))))..)))))))))..))..))..)))..((((-(((((....))))))))) ( -39.50) >DroYak_CAF1 21068 113 - 1 GCCAACGGGUUAAGGAUGUCCUGGCCUGGGGGAAACUGAAAGAUAUGUGGGCAUAGUCAGUGGGUAGCAGGGCGUUUCAAAACGAAGGUAGUGCA-UACACACAUACAUAUGUA .((..(((((((.((....)))))))))..))..........((((((((((...))).(((.((((((..((.((((.....))))))..))).-))).)))...))))))). ( -33.60) >consensus GCCAACGGGUUAAGGAUGUCCUGGCCUGGGGAAAACUGAAAGAUAUGUGGGCAUAGUCAGUGGGUAGCAGGGCGUUUCAAAACGAAGGUAGUGCA_UACAUACAUACAUACAUA (((..((..((...(((((((((((((.......(((((....((((....)))).)))))))))..)))))))))..))..))..)))......................... (-27.68 = -27.93 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:27 2006