
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,518,820 – 13,519,017 |
| Length | 197 |
| Max. P | 0.999467 |

| Location | 13,518,820 – 13,518,916 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 90.02 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -28.75 |
| Energy contribution | -29.67 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13518820 96 + 22407834 GGUAGGGGUGGCGCCACAUCAGCAGCUGCUGUUGGCCCACAGUUAACAGUGCUAACUGU-----GUUGCCUGGCACUGCGAUUUAUUAACAGUUAGCCAAA (((((..((((.((((...((((....)))).)))))))).....(((((....)))))-----.)))))((((((((..(.....)..))))..)))).. ( -35.10) >DroSec_CAF1 10045 94 + 1 GGUAG--GUGGCGCCACAUCAGCAGCUGCUGUUGGCCCACAGUUAACUGUACUAACUGU-----GUUGCCUGGCACUGCGAUUUAUUAACAGUUAGCCAAA ...((--(..((((((...((((....)))).))))..(((((((.......)))))))-----))..)))(((((((..(.....)..))))..)))... ( -32.40) >DroSim_CAF1 6669 94 + 1 GGAAG--GUGGCGCCACAUCAGCAGCUGCUGUUGGCCCACAGUUAACUGUGCUAACUGU-----GUUGCCUGGCACUGCGAUUUAUUAACAGUUAGCCAAA .((..--((((.((((...((((....)))).))))))))..))...((.(((((((((-----(((((........)))))......))))))))))).. ( -34.70) >DroEre_CAF1 10416 94 + 1 GGAAG--GUGGCGCCACAUCAGCAGCUGCUGUUGGGCCACAGUUAACUGUGCUAACUGU-----GUUGCCUGGCACUGCGAUUUAUUAACAGUUAGCCAAA .((..--(((((.(((...((((....)))).))))))))..))...((.(((((((((-----(((((........)))))......))))))))))).. ( -34.70) >DroYak_CAF1 10735 93 + 1 GGAAG--GUGGCGCCACAUCAGCAGCUGCUGUUG-CCCACAGUUAACUGUGCUAACUGU-----GUUGCCUGGCACUGCGAUUUAUUAACAGUUAGCCAAA ...((--(..((((.......(((((....))))-).(((((....)))))......))-----))..)))(((((((..(.....)..))))..)))... ( -30.10) >DroAna_CAF1 9814 97 + 1 GAUGGGGCGGGAGCCACAUCAGCAGCUGCUUCUGGCCCAG----AACUGUGCUAACUGUUUGCUGAUAUCUGGCACUGCGAUUUAUUAACAGUUAGCCAAA ..((((.(((((((..(.......)..))))))).)))).----...((.(((((((((....(((((..(.((...)).)..)))))))))))))))).. ( -33.30) >consensus GGAAG__GUGGCGCCACAUCAGCAGCUGCUGUUGGCCCACAGUUAACUGUGCUAACUGU_____GUUGCCUGGCACUGCGAUUUAUUAACAGUUAGCCAAA .......((((.((((...((((....)))).)))))))).......((.(((((((((.....(((((........)))))......))))))))))).. (-28.75 = -29.67 + 0.92)


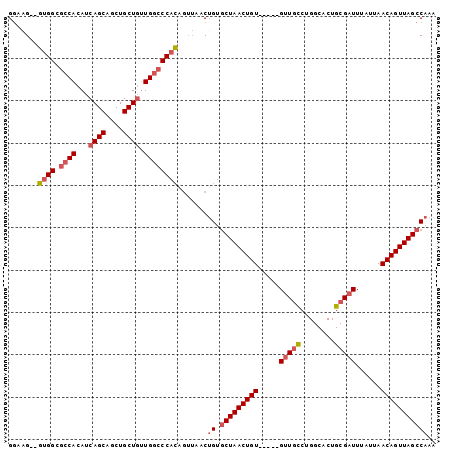
| Location | 13,518,820 – 13,518,916 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 90.02 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -26.50 |
| Energy contribution | -28.03 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13518820 96 - 22407834 UUUGGCUAACUGUUAAUAAAUCGCAGUGCCAGGCAAC-----ACAGUUAGCACUGUUAACUGUGGGCCAACAGCAGCUGCUGAUGUGGCGCCACCCCUACC ..((((..(((((.........)))))))))(((..(-----(((((((((...)))))))))).((((.((((....))))...)))))))......... ( -38.70) >DroSec_CAF1 10045 94 - 1 UUUGGCUAACUGUUAAUAAAUCGCAGUGCCAGGCAAC-----ACAGUUAGUACAGUUAACUGUGGGCCAACAGCAGCUGCUGAUGUGGCGCCAC--CUACC ..((((..(((((.........)))))))))(((..(-----((((((((.....))))))))).((((.((((....))))...)))))))..--..... ( -36.70) >DroSim_CAF1 6669 94 - 1 UUUGGCUAACUGUUAAUAAAUCGCAGUGCCAGGCAAC-----ACAGUUAGCACAGUUAACUGUGGGCCAACAGCAGCUGCUGAUGUGGCGCCAC--CUUCC ..((((..(((((.........)))))))))(((..(-----(((((((((...)))))))))).((((.((((....))))...)))))))..--..... ( -38.70) >DroEre_CAF1 10416 94 - 1 UUUGGCUAACUGUUAAUAAAUCGCAGUGCCAGGCAAC-----ACAGUUAGCACAGUUAACUGUGGCCCAACAGCAGCUGCUGAUGUGGCGCCAC--CUUCC ..((((..(((((.........)))))))))(((..(-----(((((((((...))))))))))(((((.((((....)))).)).))))))..--..... ( -39.40) >DroYak_CAF1 10735 93 - 1 UUUGGCUAACUGUUAAUAAAUCGCAGUGCCAGGCAAC-----ACAGUUAGCACAGUUAACUGUGGG-CAACAGCAGCUGCUGAUGUGGCGCCAC--CUUCC ..((((..(((((.........)))))))))(((..(-----(((((((((...)))))))))).(-((.((((....)))).)))...)))..--..... ( -34.60) >DroAna_CAF1 9814 97 - 1 UUUGGCUAACUGUUAAUAAAUCGCAGUGCCAGAUAUCAGCAAACAGUUAGCACAGUU----CUGGGCCAGAAGCAGCUGCUGAUGUGGCUCCCGCCCCAUC ((((((..(((((.........))))).(((((.....((.........)).....)----))))))))))(((....)))((((.(((....))).)))) ( -27.90) >consensus UUUGGCUAACUGUUAAUAAAUCGCAGUGCCAGGCAAC_____ACAGUUAGCACAGUUAACUGUGGGCCAACAGCAGCUGCUGAUGUGGCGCCAC__CUACC ..((((..(((((.........)))))))))(((........(((((((((...)))))))))..((((.((((....))))...)))))))......... (-26.50 = -28.03 + 1.53)


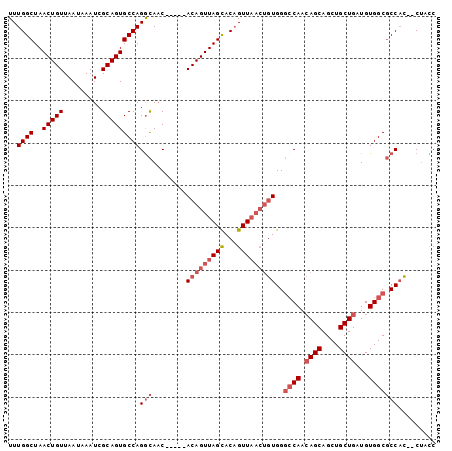
| Location | 13,518,916 – 13,519,017 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 71.43 |
| Mean single sequence MFE | -17.29 |
| Consensus MFE | -10.98 |
| Energy contribution | -11.97 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.63 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13518916 101 + 22407834 AGUCCGGAUAGCUAUAGCUACCUAC---CUACUACUCAAAUAACUUAGCUGCUACCUACAACUGCUACAAUUAUUGUAGUUAUUCUCUUACU-GGGCUCUACUGA (((((((.((((...(((((.....---.................))))))))).........((((((.....))))))..........))-)))))....... ( -19.35) >DroSec_CAF1 10139 84 + 1 AGCCCGGAUAGCU--AGCUACCUAC---CUAC---UCAA------------CUACCUACAACUGCUACAAUUAUUGUAGUUAUUCUUCUGCU-GGGCUCCACUGA (((((((.(((.(--((....))).---))).---....------------............((((((.....))))))..........))-)))))....... ( -17.70) >DroSim_CAF1 6763 84 + 1 AGCCCGGAUAGCU--AGCUACCUAC---CUAC---UCAA------------CUACCUACAACUGCUACAAUUAUUGUAGUUAUUCUUNNNNN-NNNNNNNNNNNN .....(..(((.(--((....))).---))).---.)..------------............((((((.....))))))............-............ ( -8.40) >DroEre_CAF1 10510 84 + 1 AGCCCGAAUAUCU--AUGUAGCUACUUCCUAC---CUAG------------CUACCUACAACUGCUACAAUUGUUGUAGUUAUUCUUCUACUUGGGC----CUAA .((((((......--..(((((((........---.)))------------))))(((((((..........)))))))............))))))----.... ( -23.70) >consensus AGCCCGGAUAGCU__AGCUACCUAC___CUAC___UCAA____________CUACCUACAACUGCUACAAUUAUUGUAGUUAUUCUUCUACU_GGGC____CUGA .((((((.((((....)))))).........................................((((((.....)))))).............))))........ (-10.98 = -11.97 + 1.00)
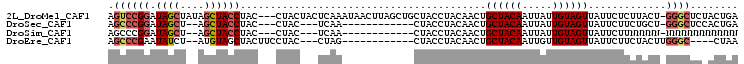

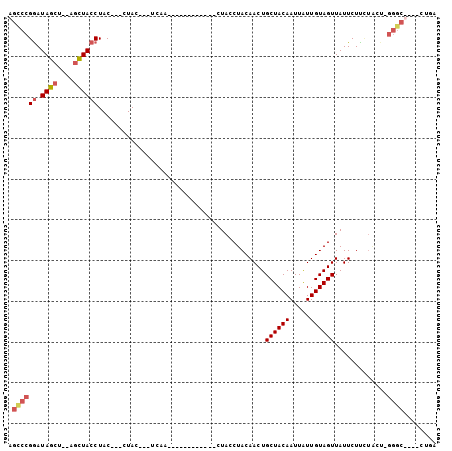
| Location | 13,518,916 – 13,519,017 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 71.43 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -11.25 |
| Energy contribution | -12.88 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13518916 101 - 22407834 UCAGUAGAGCCC-AGUAAGAGAAUAACUACAAUAAUUGUAGCAGUUGUAGGUAGCAGCUAAGUUAUUUGAGUAGUAG---GUAGGUAGCUAUAGCUAUCCGGACU ..(((...(((.-(.((...(((((((((((.....))))..((((((.....))))))..)))))))...)).).)---)).((((((....))))))...))) ( -23.00) >DroSec_CAF1 10139 84 - 1 UCAGUGGAGCCC-AGCAGAAGAAUAACUACAAUAAUUGUAGCAGUUGUAGGUAG------------UUGA---GUAG---GUAGGUAGCU--AGCUAUCCGGGCU .......(((((-..........(((((((.(((((((...)))))))..))))------------))).---((((---.(((....))--).))))..))))) ( -23.90) >DroSim_CAF1 6763 84 - 1 NNNNNNNNNNNN-NNNNNAAGAAUAACUACAAUAAUUGUAGCAGUUGUAGGUAG------------UUGA---GUAG---GUAGGUAGCU--AGCUAUCCGGGCU ............-..........(((((((.(((((((...)))))))..))))------------))).---...(---((.(((((..--..)))))...))) ( -14.80) >DroEre_CAF1 10510 84 - 1 UUAG----GCCCAAGUAGAAGAAUAACUACAACAAUUGUAGCAGUUGUAGGUAG------------CUAG---GUAGGAAGUAGCUACAU--AGAUAUUCGGGCU ...(----((((.((((.........(((((((..........)))))))((((------------(((.---........)))))))..--...)))).))))) ( -25.60) >consensus UCAG____GCCC_AGUAGAAGAAUAACUACAAUAAUUGUAGCAGUUGUAGGUAG____________UUGA___GUAG___GUAGGUAGCU__AGCUAUCCGGGCU ........((((..............(((((((..........))))))).................................((((((....)))))).)))). (-11.25 = -12.88 + 1.62)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:22 2006