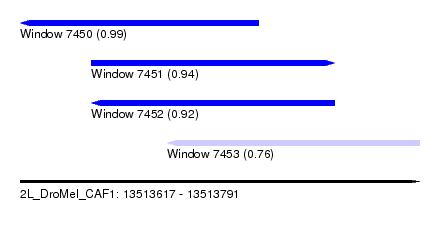
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,513,617 – 13,513,791 |
| Length | 174 |
| Max. P | 0.988801 |

| Location | 13,513,617 – 13,513,721 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.94 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -11.48 |
| Energy contribution | -12.24 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13513617 104 - 22407834 AGUCUAAUAAUAAGGCAAAAAUAUUUGCCAAACAAAAUUGAGCAGCUCGGCAAAUAUUAUU-------UUGCCGAUAAUUGUGAGAGUGUGCAGCUGUCAGAUUGAGGCCA .((((........((((((....))))))......((((..(((((((((((((......)-------)))))))....(((........))))))))..)))).)))).. ( -31.20) >DroSec_CAF1 5004 104 - 1 AGUCUAAUAAUAAGGCAAAAAUAUUUGCCAAACAAAAUUGAGCAGCUCGGUAAAUAUUAUU-------UAGCCGAUAAUUGUGAGAGUGUGCAGCUGUCAGAUUGAGGCCA .((((........((((((....))))))......((((..((((((((((.((......)-------).)))))....(((........))))))))..)))).)))).. ( -25.60) >DroEre_CAF1 4906 111 - 1 UGUCUAAUAAUAAGGUAAAAAUAUUUGCCAAACAAAAUCGAGCAGCUCGGUAAAUAUUAUUCAUAUGAUAGCCGAUAAUUGUGAGAGUGUGCAGCUGUCAGAUUGAGGCCA .((((........((((((....))))))......((((..((((((((((...((((((....)))))))))))....(((........))))))))..)))).)))).. ( -27.10) >DroYak_CAF1 5169 111 - 1 AAUGUAAUAAUAAGGUAAAAAUAUUUGCCAAACUAAAUCGAGCAGCUUGGUAAAUAUUAUUCAUAUGAUAGCCAAUAAUUGUGAGAGUGUGCAGCUGUCAGAUUGAGGCCA .............(((...((((((((((((.((.........)).)))))))))))).((((..(((((((..(((.((....)).)))...)))))))...))))))). ( -26.60) >DroAna_CAF1 3670 83 - 1 AAUG-GAUACGCAAAUAAAAAUAUUGCACUAACGAUAUCGGACA--------------------------GCCGAUAAAUGUGAGAGUGCCCAGGUGUCGGCUUAAGCUC- ....-(((((...((((....))))(((((.(((.((((((...--------------------------.))))))..)))...)))))....)))))(((....))).- ( -20.40) >consensus AGUCUAAUAAUAAGGUAAAAAUAUUUGCCAAACAAAAUCGAGCAGCUCGGUAAAUAUUAUU_______UAGCCGAUAAUUGUGAGAGUGUGCAGCUGUCAGAUUGAGGCCA .............((((((....)))))).........................................(((..(((((.(((.(((.....))).)))))))).))).. (-11.48 = -12.24 + 0.76)

| Location | 13,513,648 – 13,513,754 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.65 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -5.80 |
| Energy contribution | -7.36 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.25 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13513648 106 + 22407834 AAUUAUCGGCAA-------AAUAAUAUUUGCCGAGCUGCUCAAUUUUGUUUGGCAAAUAUUUUUGCCUUAUUAUUAGACUACCUACUUAUUUAUUGAGUGU-------GGUUUGGGCAUA .......(((((-------((..(((((((((((((...........))))))))))))))))))))......(((((((((..(((((.....)))))))-------)))))))..... ( -34.80) >DroSec_CAF1 5035 106 + 1 AAUUAUCGGCUA-------AAUAAUAUUUACCGAGCUGCUCAAUUUUGUUUGGCAAAUAUUUUUGCCUUAUUAUUAGACUACCUACUUAUUUAUUGAGUGU-------GGUUUGGGCAUU .....((((.((-------(((...)))))))))..(((((..........((((((....))))))........(((((((..(((((.....)))))))-------)))))))))).. ( -27.40) >DroEre_CAF1 4937 113 + 1 AAUUAUCGGCUAUCAUAUGAAUAAUAUUUACCGAGCUGCUCGAUUUUGUUUGGCAAAUAUUUUUACCUUAUUAUUAGACAACCUACUUAUUUAUUGAGUGU-------GGUUUGGGCACA .....((((.((..((((.....)))).))))))(.(((((((...(((((((..((((.........)))).)))))))(((((((((.....)))))).-------))))))))))). ( -21.70) >DroYak_CAF1 5200 113 + 1 AAUUAUUGGCUAUCAUAUGAAUAAUAUUUACCAAGCUGCUCGAUUUAGUUUGGCAAAUAUUUUUACCUUAUUAUUACAUUACCUACAUAUUUAUGGAGUGU-------GGUUUGGGCAUA ........((.......((((.(((((((.((((((((.......)))))))).)))))))))))((.....((((((((.((...........)))))))-------)))..))))... ( -22.80) >DroAna_CAF1 3700 93 + 1 AUUUAUCGGC--------------------------UGUCCGAUAUCGUUAGUGCAAUAUUUUUAUUUGCGUAUC-CAUUACCCAAUUAGUUAUCGAUUUUUUAUAGAGCUUUAGUAAUU ...((((((.--------------------------...))))))(((.(((.((((.........))))(((..-...)))........))).)))....................... ( -10.40) >consensus AAUUAUCGGCUA_______AAUAAUAUUUACCGAGCUGCUCGAUUUUGUUUGGCAAAUAUUUUUACCUUAUUAUUAGACUACCUACUUAUUUAUUGAGUGU_______GGUUUGGGCAUA ..............................((((((((((.(((...))).))).............................((((((.....))))))........)))))))..... ( -5.80 = -7.36 + 1.56)



| Location | 13,513,648 – 13,513,754 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.65 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -6.22 |
| Energy contribution | -7.98 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13513648 106 - 22407834 UAUGCCCAAACC-------ACACUCAAUAAAUAAGUAGGUAGUCUAAUAAUAAGGCAAAAAUAUUUGCCAAACAAAAUUGAGCAGCUCGGCAAAUAUUAUU-------UUGCCGAUAAUU ..(((.((((((-------((.............)).))).............((((((....))))))........))).)))..((((((((......)-------)))))))..... ( -24.52) >DroSec_CAF1 5035 106 - 1 AAUGCCCAAACC-------ACACUCAAUAAAUAAGUAGGUAGUCUAAUAAUAAGGCAAAAAUAUUUGCCAAACAAAAUUGAGCAGCUCGGUAAAUAUUAUU-------UAGCCGAUAAUU ..(((.((((((-------((.............)).))).............((((((....))))))........))).)))..(((((.((......)-------).)))))..... ( -18.92) >DroEre_CAF1 4937 113 - 1 UGUGCCCAAACC-------ACACUCAAUAAAUAAGUAGGUUGUCUAAUAAUAAGGUAAAAAUAUUUGCCAAACAAAAUCGAGCAGCUCGGUAAAUAUUAUUCAUAUGAUAGCCGAUAAUU ((((.......)-------)))...............(((((((.(((((((.((((((....)))))).......((((((...))))))...))))))).....)))))))....... ( -22.70) >DroYak_CAF1 5200 113 - 1 UAUGCCCAAACC-------ACACUCCAUAAAUAUGUAGGUAAUGUAAUAAUAAGGUAAAAAUAUUUGCCAAACUAAAUCGAGCAGCUUGGUAAAUAUUAUUCAUAUGAUAGCCAAUAAUU ..(((.(..(((-------(((...........))).))).............((((((....))))))..........).)))..(((((...((((((....)))))))))))..... ( -16.30) >DroAna_CAF1 3700 93 - 1 AAUUACUAAAGCUCUAUAAAAAAUCGAUAACUAAUUGGGUAAUG-GAUACGCAAAUAAAAAUAUUGCACUAACGAUAUCGGACA--------------------------GCCGAUAAAU ......................(((.((.(((.....))).)).-)))..((((.........))))........((((((...--------------------------.))))))... ( -12.80) >consensus UAUGCCCAAACC_______ACACUCAAUAAAUAAGUAGGUAGUCUAAUAAUAAGGUAAAAAUAUUUGCCAAACAAAAUCGAGCAGCUCGGUAAAUAUUAUU_______UAGCCGAUAAUU .....................................(((.............((((((....)))))).......((((((...))))))...................)))....... ( -6.22 = -7.98 + 1.76)


| Location | 13,513,681 – 13,513,791 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.82 |
| Mean single sequence MFE | -21.53 |
| Consensus MFE | -9.02 |
| Energy contribution | -11.46 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13513681 110 - 22407834 --UGGGGGUGACACAACUCCCCGAUAUAAGGUACU-CUACUAUGCCCAAACC-------ACACUCAAUAAAUAAGUAGGUAGUCUAAUAAUAAGGCAAAAAUAUUUGCCAAACAAAAUUG --.((((((......)).))))..(((.((((((.-(((((...........-------..............)))))))).))).)))....((((((....))))))........... ( -22.71) >DroSec_CAF1 5068 110 - 1 --UGGGAGUGACACACCUCCCCGAUAUUAGGUACC-CUACAAUGCCCAAACC-------ACACUCAAUAAAUAAGUAGGUAGUCUAAUAAUAAGGCAAAAAUAUUUGCCAAACAAAAUUG --.(((((........)))))...(((((((((((-.(((............-------...............))))))).)))))))....((((((....))))))........... ( -23.43) >DroEre_CAF1 4977 110 - 1 --UGGGAGUGAUACAGCUCCCUGAUAUUGGGUACC-CUACUGUGCCCAAACC-------ACACUCAAUAAAUAAGUAGGUUGUCUAAUAAUAAGGUAAAAAUAUUUGCCAAACAAAAUCG --.((((((......)))))).((((((((((((.-.....))))))))(((-------((.............)).))))))).........((((((....))))))........... ( -29.02) >DroYak_CAF1 5240 111 - 1 --UGGCAGUGACACACCUCCCCAAUAUUGGGUACCCCUACUAUGCCCAAACC-------ACACUCCAUAAAUAUGUAGGUAAUGUAAUAAUAAGGUAAAAAUAUUUGCCAAACUAAAUCG --((((((......((((........(((((((.........)))))))(((-------(((...........))).)))............))))........)))))).......... ( -19.64) >DroAna_CAF1 3714 112 - 1 AAUGACAGUAUAGGAAUUCCUC------UAGUACC-CCACAAUUACUAAAGCUCUAUAAAAAAUCGAUAACUAAUUGGGUAAUG-GAUACGCAAAUAAAAAUAUUGCACUAACGAUAUCG ..((.((((((.((....))((------((.((((-(...(((((..........................)))))))))).))-)).............))))))))............ ( -12.87) >consensus __UGGGAGUGACACAACUCCCCGAUAUUAGGUACC_CUACUAUGCCCAAACC_______ACACUCAAUAAAUAAGUAGGUAGUCUAAUAAUAAGGUAAAAAUAUUUGCCAAACAAAAUCG ...((((((......))))))...(((((((((((..(((..................................))))))).)))))))....((((((....))))))........... ( -9.02 = -11.46 + 2.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:17 2006