
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,498,664 – 13,498,791 |
| Length | 127 |
| Max. P | 0.844508 |

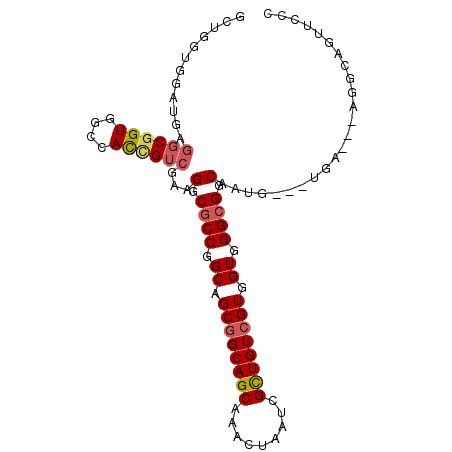
| Location | 13,498,664 – 13,498,758 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -30.48 |
| Energy contribution | -30.73 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
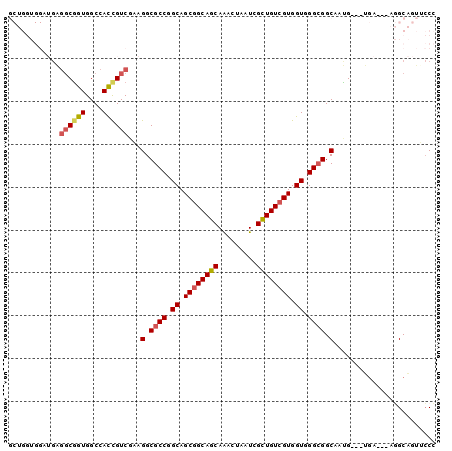
>2L_DroMel_CAF1 13498664 94 - 22407834 GCUGGUGGAUGAGGCGGUGGCCACCGUCGAAGGCGCCGGCAGCGGCAGCAGACUAAUCGCUGUCGUGGUGGGCGGCAAUG---UGA---AGGCAGUUCCC (((..((.((..((((((....))))))...(.((((.((.((((((((.((....)))))))))).)).)))).).)).---)).---.)))....... ( -40.70) >DroSec_CAF1 97486 94 - 1 GCUGGUGGAUGAGGCGGUGGCCACCGUCGAAGGCGCCGGCAGCGGCAGCAAACUAAUCGUUGUCGUGGUGGGCGGCAAUG---UGA---AGGCAGUUCCC (((..((.((..((((((....))))))...(.((((.((.((((((((.........)))))))).)).)))).).)).---)).---.)))....... ( -37.90) >DroSim_CAF1 99022 94 - 1 GCUGGUGGAUGAGGCGGUGGCCACCGUCGAAGGCGCCGGCAGCGGCAGCAAACUAAUCGCUGUCGUGGUGGGCGGCAAUG---UGA---AGGCAGUUCCC (((..((.((..((((((....))))))...(.((((.((.((((((((.........)))))))).)).)))).).)).---)).---.)))....... ( -40.30) >DroEre_CAF1 101994 94 - 1 GCUGGUGGAUGAGGCGGUGGCCACCGUCGAAGGCGCCGGCAGCGGCAGCAAGCUAAUUGCUGUCGUGGUGGGCGGCAUUG---UGA---AGGCAGUGCCC (((((((..(..((((((....))))))..)..))))))).((((((((((.....)))))))))).......(((((((---(..---..)))))))). ( -49.50) >DroAna_CAF1 161805 87 - 1 ----------GGGGCGGUGGCCAUUGUCGGCGGCGCCGGCAGCGGCAGCAAACUGAUCGUUGUGGUGGUGGGAGGCUGCG---UUCCUGCUGCCGUUCCC ----------(((((((..(((.((((((((...)))))))).)(((((...((.((((......)))).))..))))).---.....))..))))))). ( -42.20) >DroPer_CAF1 118659 83 - 1 --------------CCGUGGCCAUUGUCGGCGGCGCCGGCAGCGGCAGCAGGCUGAUCGUUGUCGUGGUGGGCGGCAGGGCCGCGA---AUGCGGAUGCG --------------.(((((((.((((((.(.((..((((((((((((....))).)))))))))..)).).))))))))))))).---..((....)). ( -46.00) >consensus GCUGGUGGAUGAGGCGGUGGCCACCGUCGAAGGCGCCGGCAGCGGCAGCAAACUAAUCGCUGUCGUGGUGGGCGGCAAUG___UGA___AGGCAGUUCCC ............((((((....))))))...(.((((.((.((((((((.........)))))))).)).)))).)........................ (-30.48 = -30.73 + 0.25)

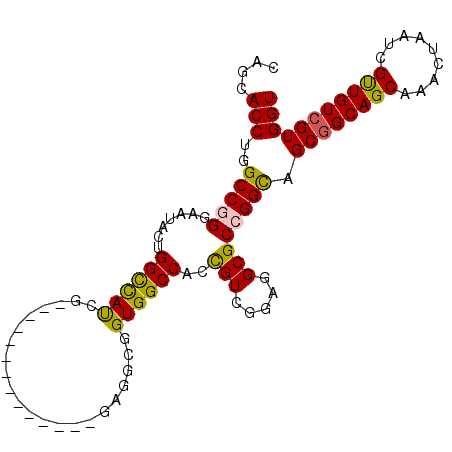
| Location | 13,498,689 – 13,498,791 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -43.67 |
| Consensus MFE | -29.34 |
| Energy contribution | -28.62 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13498689 102 - 22407834 CAGCACCUGGCCGGGAAUACUGGCUAUCGUUGUGCUGGUGGAUGAGGCGGUGGCCACCGUCGAAGGCGCCGGCAGCGGCAGCAGACUAAUCGCUGUCGUGGU ((((...(((((((.....)))))))..))))((((((((..(..((((((....))))))..)..))))))))((((((((.((....))))))))))... ( -49.10) >DroPse_CAF1 141217 84 - 1 CGGCACCUGGCCAGGAAUACUGGUCAUCG------------------CCGUGGCCAUUGUCGGCGGCGCCGGUAGCGGCAGCAGGCUGAUCGUUGUCGUGGU ((((...(((((((.....)))))))..(------------------(((((((....))).)))))))))...((((((((.........))))))))... ( -38.50) >DroSec_CAF1 97511 102 - 1 CAGCACCUGGCCGGGAAUACAGGCUAUCGUUGUGCUGGUGGAUGAGGCGGUGGCCACCGUCGAAGGCGCCGGCAGCGGCAGCAAACUAAUCGUUGUCGUGGU ((((...(((((.(.....).)))))..))))((((((((..(..((((((....))))))..)..))))))))((((((((.........))))))))... ( -42.40) >DroYak_CAF1 99633 102 - 1 CAGCACCUGGCCGGGAAUACUGGCUAUCGUUGUGCUGGUGGAUGAGGCGGUGGCCACCGUCGGAGGCGCCGGCAGCGGCAGCAAACUAAUUGCUGUCGUGGU ((((...(((((((.....)))))))..))))((((((((..(..((((((....))))))..)..))))))))((((((((((.....))))))))))... ( -50.90) >DroAna_CAF1 161833 87 - 1 CAGGACCUGGCCGGGAAUGCUGGCCACA---------------GGGGCGGUGGCCAUUGUCGGCGGCGCCGGCAGCGGCAGCAAACUGAUCGUUGUGGUGGU ....(((.((((((.....))))))(((---------------(.((((((.(((.((((((((...)))))))).))).....)))).)).)))))))... ( -39.20) >DroPer_CAF1 118687 84 - 1 CGGCACCUGGCCAGGAAUACUGGUCAUCG------------------CCGUGGCCAUUGUCGGCGGCGCCGGCAGCGGCAGCAGGCUGAUCGUUGUCGUGGU .(((...(((((((.....)))))))..(------------------(((((((....))).))))))))(((((((((((....))).))))))))..... ( -41.90) >consensus CAGCACCUGGCCGGGAAUACUGGCCAUCG______________GAGGCGGUGGCCACCGUCGGAGGCGCCGGCAGCGGCAGCAAACUAAUCGUUGUCGUGGU ....(((..(((((.......((((((......................))))))..(((.....)))))))).((((((((.........))))))))))) (-29.34 = -28.62 + -0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:04 2006