| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,370,542 – 1,370,650 |
| Length | 108 |
| Max. P | 0.716461 |

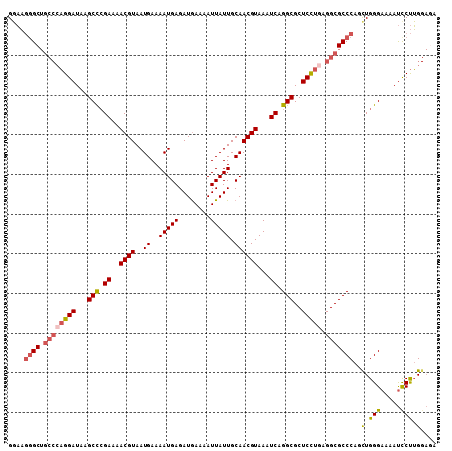
| Location | 1,370,542 – 1,370,650 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.19 |
| Mean single sequence MFE | -31.71 |
| Consensus MFE | -26.48 |
| Energy contribution | -27.12 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1370542 108 + 22407834 GGAAGGGCUGCCCAGGAUAAGCCCGAAAACGUAAUGAAAAUGAGAUAAAAAUUAUUGCAACGUAAAUCAGGCGCUCUUGAGGCGCCCAGCUGGGAAUAUUCUUGGAGA ....((((.((((((((...(((.((..((((..((..(((((........))))).))))))...)).)))..))))).)))))))..(..(((....)))..)... ( -33.40) >DroSec_CAF1 2196 108 + 1 GGAAGGGCUGCCCAGGAUAAGCCCGAAAACGUAAUGAAAAUGAGAUGAAAAUUAUUGCAACGUAAAUCAGGCGCUCCUUCGGCGCCCAGCUGGGAAAAUCCUUGGAGA ....((((.(((.((((...(((.((..((((..((..(((((........))))).))))))...)).)))..))))..)))))))..(..((......))..)... ( -30.80) >DroSim_CAF1 2181 108 + 1 GGAAGGGCUGCCCAGGAUAAGCCCGAAAACGUAAUGAAAAUGAGAUGAAAAUUAUUGCAACGUAAAUCAGGCGCUCCUGCGGCGCCCAGCUGGGAAAAUCCUUGGAGA ....((((.((((((((...(((.((..((((..((..(((((........))))).))))))...)).)))..))))).)))))))..(..((......))..)... ( -35.10) >DroEre_CAF1 2062 107 + 1 GGAAG-GCUGCCCAGGAUAAGCCCGAAAACGUAAUGAAAAUGAGAUGAAAAUUAUUGCAACGUAAAUCAGGCGCUCCUGAGGCGCCCAGCUGGGAAAAUCCUUGGAUG ....(-((.((((((((...(((.((..((((..((..(((((........))))).))))))...)).)))..))))).))))))...(..((......))..)... ( -30.90) >DroYak_CAF1 2092 108 + 1 GGAAGGGCUGCCCAGGAUAAGCUCGAAAACGUAAUGAAAAUGAGAUGAAAAUUAUUGCAACGUAAAUCAGGCGCUCCUGAGGCGCCCAGUUGGGAAAAUCCUUGGAGA ....((((.((((((((...((..((..((((..((..(((((........))))).))))))...))..))..))))).)))))))....(((.....)))...... ( -32.00) >DroAna_CAF1 2582 99 + 1 GCUCCGGC-----CGGAUAAGCCAGAGAACGUAAUGAAAAUGAGAUGAAAAUUAUUGCAACGUAAAUCAGGCGCUCCUGAGGCGCCCAGCUGGGAAUCCCCCGG---- ...((((.-----.((((...((((.....(((((((..............)))))))...........((((((.....))))))...))))..)))).))))---- ( -28.04) >consensus GGAAGGGCUGCCCAGGAUAAGCCCGAAAACGUAAUGAAAAUGAGAUGAAAAUUAUUGCAACGUAAAUCAGGCGCUCCUGAGGCGCCCAGCUGGGAAAAUCCUUGGAGA ....((((.((((((((...(((.((..((((..((..(((((........))))).))))))...)).)))..))))).)))))))..(.(((.....))).).... (-26.48 = -27.12 + 0.64)


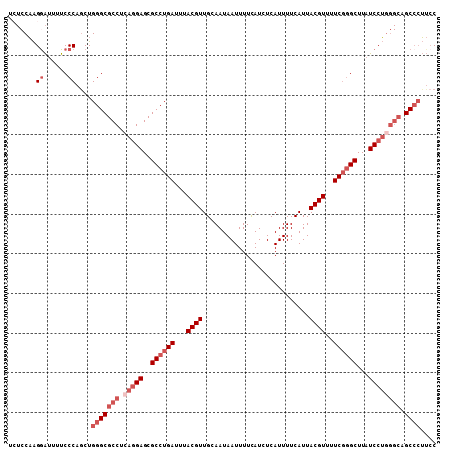
| Location | 1,370,542 – 1,370,650 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.19 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -24.93 |
| Energy contribution | -26.93 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1370542 108 - 22407834 UCUCCAAGAAUAUUCCCAGCUGGGCGCCUCAAGAGCGCCUGAUUUACGUUGCAAUAAUUUUUAUCUCAUUUUCAUUACGUUUUCGGGCUUAUCCUGGGCAGCCCUUCC .....................(((((((.((.((..((((((...((((...........................))))..))))))...)).))))).)))).... ( -27.83) >DroSec_CAF1 2196 108 - 1 UCUCCAAGGAUUUUCCCAGCUGGGCGCCGAAGGAGCGCCUGAUUUACGUUGCAAUAAUUUUCAUCUCAUUUUCAUUACGUUUUCGGGCUUAUCCUGGGCAGCCCUUCC .......((......))....(((((((..((((..((((((...((((...........................))))..))))))...)))).))).)))).... ( -32.43) >DroSim_CAF1 2181 108 - 1 UCUCCAAGGAUUUUCCCAGCUGGGCGCCGCAGGAGCGCCUGAUUUACGUUGCAAUAAUUUUCAUCUCAUUUUCAUUACGUUUUCGGGCUUAUCCUGGGCAGCCCUUCC .......((......))....(((((((.(((((..((((((...((((...........................))))..))))))...)))))))).)))).... ( -35.63) >DroEre_CAF1 2062 107 - 1 CAUCCAAGGAUUUUCCCAGCUGGGCGCCUCAGGAGCGCCUGAUUUACGUUGCAAUAAUUUUCAUCUCAUUUUCAUUACGUUUUCGGGCUUAUCCUGGGCAGC-CUUCC .......((......))....(((((((.(((((..((((((...((((...........................))))..))))))...)))))))).))-))... ( -33.03) >DroYak_CAF1 2092 108 - 1 UCUCCAAGGAUUUUCCCAACUGGGCGCCUCAGGAGCGCCUGAUUUACGUUGCAAUAAUUUUCAUCUCAUUUUCAUUACGUUUUCGAGCUUAUCCUGGGCAGCCCUUCC .......((......))....(((((((.(((((..((.(((...((((...........................))))..))).))...)))))))).)))).... ( -29.13) >DroAna_CAF1 2582 99 - 1 ----CCGGGGGAUUCCCAGCUGGGCGCCUCAGGAGCGCCUGAUUUACGUUGCAAUAAUUUUCAUCUCAUUUUCAUUACGUUCUCUGGCUUAUCCG-----GCCGGAGC ----.(((((....)))....(((((((....).))))))......)).................................((((((((.....)-----))))))). ( -26.90) >consensus UCUCCAAGGAUUUUCCCAGCUGGGCGCCUCAGGAGCGCCUGAUUUACGUUGCAAUAAUUUUCAUCUCAUUUUCAUUACGUUUUCGGGCUUAUCCUGGGCAGCCCUUCC .......((......))....(((((((.(((((..((((((...((((...........................))))..))))))...)))))))).)))).... (-24.93 = -26.93 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:56 2006