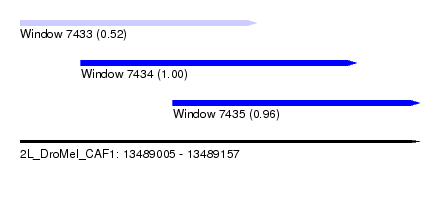
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,489,005 – 13,489,157 |
| Length | 152 |
| Max. P | 0.999306 |

| Location | 13,489,005 – 13,489,095 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -11.90 |
| Energy contribution | -11.74 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13489005 90 + 22407834 UAAA--------GUAUUAAAGUGUUAUUUUGCCAACAAAAUCAAACAAAUUUGGC--ACGUAUAACUA-----UGCAUACUUUUGGGGCACACUUAGCCUUGAAU ..((--------((((......(((((..((((((...............)))))--)...)))))..-----...))))))(..((((.......))))..).. ( -19.76) >DroSec_CAF1 87764 97 + 1 UAAA--------GUAAUAAAGUGUUAUUUUGCCAACGAAAUCAAACAAAUUUGGCGCACCUAUUACUAUGGUAUGCAUACUUUUGGGGCAGACUUAGCCUUGAAU ...(--------((((((..((((......(((((...............))))))))).)))))))..((((....))))((..((((.......))))..)). ( -19.76) >DroSim_CAF1 89342 90 + 1 UAAA--------GUAUUAAAGUGUUAUUUUGCCAACGAAAUCGAACAAAUUUGGC--ACGCAUUACUA-----UGCAUACUUUUGGGGCAGACUUAGCCUUGAAU ..((--------((((....((((((.((((....((....))..))))..))))--))((((....)-----)))))))))(..((((.......))))..).. ( -21.70) >DroEre_CAF1 92513 90 + 1 ------AUAAAAGUAUUAAAGUGAUAUUUUGCCAACGAAAUCAACCAAAUUUGGC----GUAUUACUA-----UGCAUACUUUUGGGGCAGACUUAGCCUUGAAU ------.(((((((((((.((((((((...(((((...............)))))----)))))))).-----)).)))))))))((((.......))))..... ( -21.86) >DroYak_CAF1 89800 88 + 1 AAAAAAAUAAAAGGAUUGAA----------GCCGACGAAAUCAAACAAAUUUGGC--ACGUAUUACUA-----UGCAUACUUUUGGGGCAGACCUAGCCUUGAAU ....................----------(((((...............)))))--..((((....)-----))).....((..((((.......))))..)). ( -14.86) >consensus UAAA________GUAUUAAAGUGUUAUUUUGCCAACGAAAUCAAACAAAUUUGGC__ACGUAUUACUA_____UGCAUACUUUUGGGGCAGACUUAGCCUUGAAU ..............................(((((...............)))))..........................((..((((.......))))..)). (-11.90 = -11.74 + -0.16)

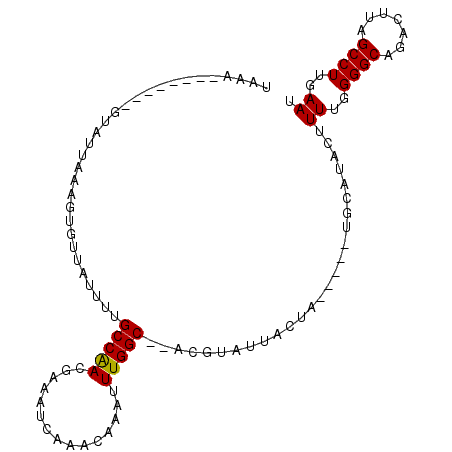
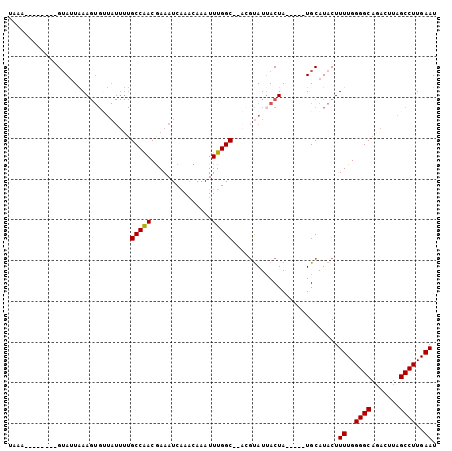
| Location | 13,489,028 – 13,489,133 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -22.95 |
| Energy contribution | -23.15 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.999306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13489028 105 + 22407834 CCAACAAAAUCAAACAAAUUUGGC--ACGUAUAACUA-----UGCAUACUUUUGGGGCACACUUAGCCUUGAAUUAUUAUUAUAAAGGCCCUUAAAAGUAUGUAUAAAGCCA ....................((((--..(.....)((-----(((((((((((((((........(((((.(((....)))...))))))))))))))))))))))..)))) ( -28.60) >DroSec_CAF1 87787 112 + 1 CCAACGAAAUCAAACAAAUUUGGCGCACCUAUUACUAUGGUAUGCAUACUUUUGGGGCAGACUUAGCCUUGAAUUAUUAUUAUAAAAGCCCUUAAAAGUAUGUAUAAAGCCA ....................((((....((((....))))((((((((((((((((((...(........)..((((....))))..)))).))))))))))))))..)))) ( -27.70) >DroSim_CAF1 89365 105 + 1 CCAACGAAAUCGAACAAAUUUGGC--ACGCAUUACUA-----UGCAUACUUUUGGGGCAGACUUAGCCUUGAAUUAUUAUUACAAAGGCCCUUAAAAGUAUGUAUAAAGCCA ....((....))........((((--..(.....)((-----(((((((((((((((........(((((.(((....)))...))))))))))))))))))))))..)))) ( -28.30) >DroEre_CAF1 92538 103 + 1 CCAACGAAAUCAACCAAAUUUGGC----GUAUUACUA-----UGCAUACUUUUGGGGCAGACUUAGCCUUGAAUUACAAUUAUAAAGGCCCUUAAAAGUAUGUAUAAAGCCA ....................((((----.......((-----(((((((((((((((........(((((.(((....)))...))))))))))))))))))))))..)))) ( -28.10) >DroYak_CAF1 89821 105 + 1 CCGACGAAAUCAAACAAAUUUGGC--ACGUAUUACUA-----UGCAUACUUUUGGGGCAGACCUAGCCUUGAAUUAUAAUUAAAAAGGCCCUUAAAAGUAUGUAUAAAGCCA ....................((((--..(.....)((-----(((((((((((((((........(((((.(((....)))...))))))))))))))))))))))..)))) ( -28.10) >consensus CCAACGAAAUCAAACAAAUUUGGC__ACGUAUUACUA_____UGCAUACUUUUGGGGCAGACUUAGCCUUGAAUUAUUAUUAUAAAGGCCCUUAAAAGUAUGUAUAAAGCCA ....................((((..................(((((((((((((((........(((((.(((....)))...))))))))))))))))))))....)))) (-22.95 = -23.15 + 0.20)

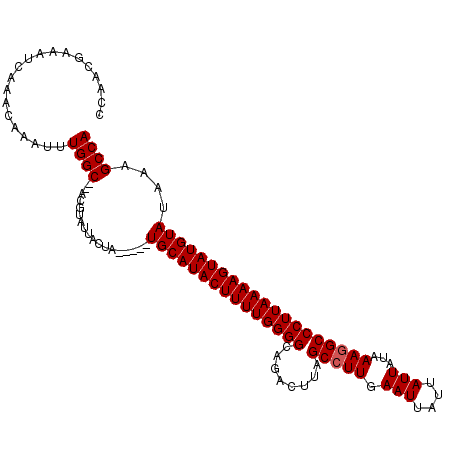
| Location | 13,489,063 – 13,489,157 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -19.14 |
| Energy contribution | -20.03 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13489063 94 + 22407834 --UGCAUACUUUUGGGGCACACUUAGCCUUGA--AU----UAUUAUUAUAAAGGCCCUUAAAAGUAUGUAUAAAGCCAGU---ACCG------AUAGGUUACACAUUAA--AA --(((((((((((((((........(((((.(--((----....)))...))))))))))))))))))))........((---(((.------...)).))).......--.. ( -26.00) >DroSec_CAF1 87827 96 + 1 UAUGCAUACUUUUGGGGCAGACUUAGCCUUGA--AU----UAUUAUUAUAAAAGCCCUUAAAAGUAUGUAUAAAGCCAGU---ACCG------AUAGGUUACACAUUAA--AA ((((((((((((((((((...(........).--.(----(((....))))..)))).))))))))))))))......((---(((.------...)).))).......--.. ( -25.10) >DroSim_CAF1 89400 94 + 1 --UGCAUACUUUUGGGGCAGACUUAGCCUUGA--AU----UAUUAUUACAAAGGCCCUUAAAAGUAUGUAUAAAGCCAGU---ACCG------AUAGGUUACACAUUAA--AA --(((((((((((((((........(((((.(--((----....)))...))))))))))))))))))))........((---(((.------...)).))).......--.. ( -26.00) >DroEre_CAF1 92571 102 + 1 --UGCAUACUUUUGGGGCAGACUUAGCCUUGA--AU----UACAAUUAUAAAGGCCCUUAAAAGUAUGUAUAAAGCCAGU---ACCGAAAAUGCUAGGUUACACAUUAAAAAA --(((((((((((((((........(((((.(--((----....)))...))))))))))))))))))))...(((((((---(.......)))).))))............. ( -26.80) >DroYak_CAF1 89856 102 + 1 --UGCAUACUUUUGGGGCAGACCUAGCCUUGA--AU----UAUAAUUAAAAAGGCCCUUAAAAGUAUGUAUAAAGCCAGU---ACCGGAAAUGAUAGGUUACACACUAAAAAA --(((((((((((((((........(((((.(--((----....)))...))))))))))))))))))))........((---(((..........)).)))........... ( -24.60) >DroAna_CAF1 152011 103 + 1 --UGCAUACUUUUGGUGAAAAAUUGGGCUUAAGAAUGUCGUA-UGUCUCAAAGACCCUGAAAAGUAUGCUAAAAAAAAGGUCAAUCG------UUAGG-GAUACAUUACAAAA --.((((((((((((..........(((........)))...-.(((.....)))))..))))))))))..............(((.------.....-)))........... ( -16.20) >consensus __UGCAUACUUUUGGGGCAGACUUAGCCUUGA__AU____UAUUAUUAUAAAGGCCCUUAAAAGUAUGUAUAAAGCCAGU___ACCG______AUAGGUUACACAUUAA__AA ..(((((((((((((((........(((((....................))))))))))))))))))))........((...(((..........)))...))......... (-19.14 = -20.03 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:00 2006