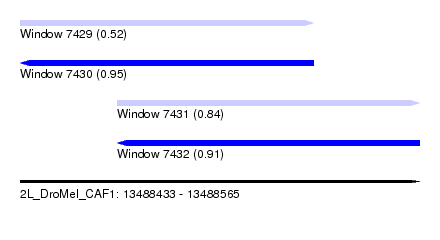
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,488,433 – 13,488,565 |
| Length | 132 |
| Max. P | 0.947478 |

| Location | 13,488,433 – 13,488,530 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -15.95 |
| Energy contribution | -15.23 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
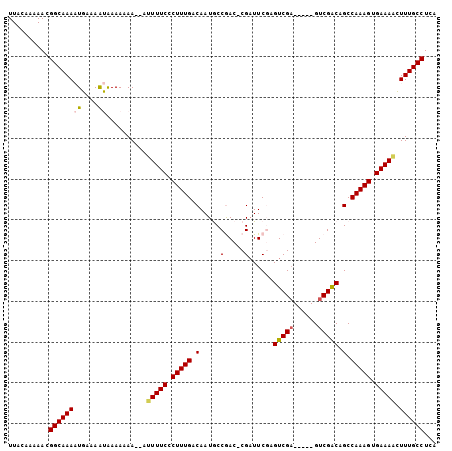
>2L_DroMel_CAF1 13488433 97 + 22407834 UUACAAAAACGGCAAAAUGCAAAUAAAAAAA--AUUUUCCCUUUGACAAUGCCGAC-CGAUUCGCGUCGA-----GUCGACAGCCAAAGUGAAACUUUUGCCUUA ..........((((((((....)).......--..((((.(((((....((.((((-((((....)))).-----)))).))..))))).))))..))))))... ( -21.60) >DroPse_CAF1 130476 103 + 1 UUACAAAAACGGCAAACUGAAAACGAAACAA--AUUUUCACUUUGACAAUACCAACCCGAUUCGAGUCGAGUCGCGUCGGCUGCCAAAGCGAAAACUUUGCCUCA ..........((((((.(....)........--.(((((.(((((.((...((.((.(((((((...))))))).)).)).)).))))).))))).))))))... ( -24.90) >DroEre_CAF1 91933 97 + 1 UUACAAAAACGGCAAAAUGCAAAUAAAAAAA--UGUUUCCCUUUGACAAUGCCGAC-CGAUUCGCGUCGA-----GUCGACAGCCAAAGUGAAACUUUUGCCUUA ..........(((((((..............--.(((((.(((((....((.((((-((((....)))).-----)))).))..))))).))))))))))))... ( -25.36) >DroYak_CAF1 89222 99 + 1 UUACAAAAACGGCAAAAUGCAAAUAAAAAAAAUUGUUUCCCUUUGACAAUGCCGAC-CGAUUCGCGUCGA-----GUCGACAGCCAAAGUGAAACUUUUGCCUUA ..........(((((((.................(((((.(((((....((.((((-((((....)))).-----)))).))..))))).))))))))))))... ( -25.22) >DroAna_CAF1 151385 97 + 1 UUACAAAAACGGCAAAAUGAAAGUGAAACAA--AUUUUCCCUUUGACAAUGCCGAC-CGUUUCUAGUCGU-----GUCGGCUGCCAAAGUGAAAACUUUGCCUCA ..........((((((((....)).......--.(((((.(((((.((..((((((-((........)).-----)))))))).))))).))))).))))))... ( -27.80) >DroPer_CAF1 106835 103 + 1 UUACAAAAACGGCAAACUGAAAACGAAACAA--AUUUUCACUUUGACAAUACCAACCCGAUUCCAGUCGAGUCGCGUCGGCUGCCAAAGCGAAAACUUUGCCUCA ..........((((((.(....)........--.(((((.(((((.((...((.((.((((((.....)))))).)).)).)).))))).))))).))))))... ( -23.50) >consensus UUACAAAAACGGCAAAAUGAAAAUAAAAAAA__AUUUUCCCUUUGACAAUGCCGAC_CGAUUCGAGUCGA_____GUCGACAGCCAAAGUGAAAACUUUGCCUCA ..........((((((.((....)).........(((((.(((((.(.....(.....)......(((((......))))).).))))).))))).))))))... (-15.95 = -15.23 + -0.72)

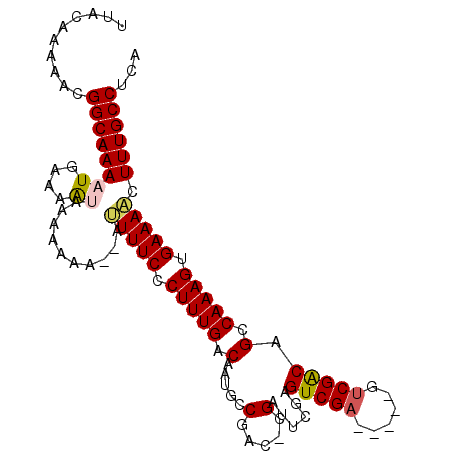
| Location | 13,488,433 – 13,488,530 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -26.41 |
| Energy contribution | -25.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.10 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13488433 97 - 22407834 UAAGGCAAAAGUUUCACUUUGGCUGUCGAC-----UCGACGCGAAUCG-GUCGGCAUUGUCAAAGGGAAAAU--UUUUUUUAUUUGCAUUUUGCCGUUUUUGUAA ...(((((((.((((.((((((((((((((-----.(((......)))-)))))))..))))))).))))..--..............))))))).......... ( -30.73) >DroPse_CAF1 130476 103 - 1 UGAGGCAAAGUUUUCGCUUUGGCAGCCGACGCGACUCGACUCGAAUCGGGUUGGUAUUGUCAAAGUGAAAAU--UUGUUUCGUUUUCAGUUUGCCGUUUUUGUAA .((((((((.((((((((((((((((((((.(((.(((...))).))).))))))..)))))))))))))))--)))))))........................ ( -40.80) >DroEre_CAF1 91933 97 - 1 UAAGGCAAAAGUUUCACUUUGGCUGUCGAC-----UCGACGCGAAUCG-GUCGGCAUUGUCAAAGGGAAACA--UUUUUUUAUUUGCAUUUUGCCGUUUUUGUAA ...((((((((((((.((((((((((((((-----.(((......)))-)))))))..))))))).)))))(--(......)).....))))))).......... ( -32.50) >DroYak_CAF1 89222 99 - 1 UAAGGCAAAAGUUUCACUUUGGCUGUCGAC-----UCGACGCGAAUCG-GUCGGCAUUGUCAAAGGGAAACAAUUUUUUUUAUUUGCAUUUUGCCGUUUUUGUAA ...((((((((((((.((((((((((((((-----.(((......)))-)))))))..))))))).)))))((......)).......))))))).......... ( -33.20) >DroAna_CAF1 151385 97 - 1 UGAGGCAAAGUUUUCACUUUGGCAGCCGAC-----ACGACUAGAAACG-GUCGGCAUUGUCAAAGGGAAAAU--UUGUUUCACUUUCAUUUUGCCGUUUUUGUAA (((((((((.(((((.((((((((((((((-----.((........))-))))))..))))))))))))).)--))))))))....................... ( -34.30) >DroPer_CAF1 106835 103 - 1 UGAGGCAAAGUUUUCGCUUUGGCAGCCGACGCGACUCGACUGGAAUCGGGUUGGUAUUGUCAAAGUGAAAAU--UUGUUUCGUUUUCAGUUUGCCGUUUUUGUAA .((((((((.((((((((((((((((((((.(((.((.....)).))).))))))..)))))))))))))))--)))))))........................ ( -40.10) >consensus UAAGGCAAAAGUUUCACUUUGGCAGCCGAC_____UCGACGCGAAUCG_GUCGGCAUUGUCAAAGGGAAAAU__UUGUUUCAUUUGCAUUUUGCCGUUUUUGUAA ...((((((((((((.(((((((.((((((.....((.....)).....))))))...))))))).))))).................))))))).......... (-26.41 = -25.97 + -0.44)

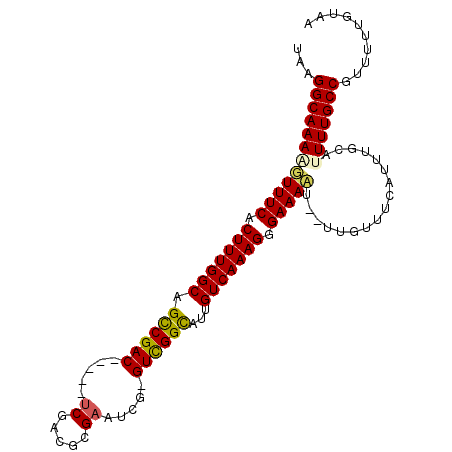
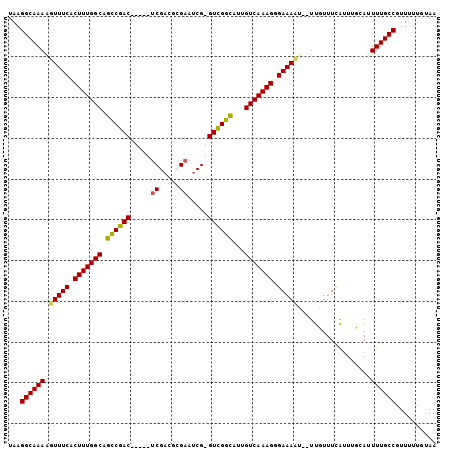
| Location | 13,488,465 – 13,488,565 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -15.23 |
| Energy contribution | -16.30 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13488465 100 + 22407834 UUUUCCCUUUGACAAUGCCGAC-CGAUUCGCGUCGA-----GUCGACAGCCAAAGUGAAACUUUUGCCUUAAAUUGAAAUGCACAUUUUAAUUUCGGCCCGAAACC---- .((((.(((((....((.((((-((((....)))).-----)))).))..))))).)))).....(((..((((((((((....)))))))))).)))........---- ( -25.30) >DroPse_CAF1 130508 106 + 1 UUUUCACUUUGACAAUACCAACCCGAUUCGAGUCGAGUCGCGUCGGCUGCCAAAGCGAAAACUUUGCCUCAAAUUUAAAUGCACAUUUUAAUUUCGGCCCGAAACC---- (((((.(((((.((...((.((.(((((((...))))))).)).)).)).))))).)))))....(((..(((((.((((....)))).))))).)))........---- ( -24.90) >DroSec_CAF1 87221 100 + 1 UUUUCCCUUUGACAAUGCCGAC-CGAUUCGCGUCGA-----GUCGACAGCCAAAGUGAAACUUUUGCCUUAAAUUGAAAUGCACAUUUUAAUUUCGGCCCAAAACC---- .((((.(((((....((.((((-((((....)))).-----)))).))..))))).)))).....(((..((((((((((....)))))))))).)))........---- ( -25.30) >DroWil_CAF1 138682 91 + 1 UGUUCACUUUGACAAUGCCCCC-CGAC------------------AAUGCCAAAGUGAAAACUUUGCCUCGAAUUGAAAUGAAAAUUUUAAUUUCGGCUCAAAACCGUUU ..(((((((((.((.((.....-...)------------------).)).)))))))))......(((..((((((((((....)))))))))).)))............ ( -23.60) >DroAna_CAF1 151417 100 + 1 UUUUCCCUUUGACAAUGCCGAC-CGUUUCUAGUCGU-----GUCGGCUGCCAAAGUGAAAACUUUGCCUCAAAUUGAAAUGCACAUUUUAAUUUCGGCCCGAAACC---- (((((.(((((.((..((((((-((........)).-----)))))))).))))).)))))....(((..((((((((((....)))))))))).)))........---- ( -29.90) >DroPer_CAF1 106867 106 + 1 UUUUCACUUUGACAAUACCAACCCGAUUCCAGUCGAGUCGCGUCGGCUGCCAAAGCGAAAACUUUGCCUCAAAUUUAAAUGCACAUUUUAAUUUCGGCCCGAAACC---- (((((.(((((.((...((.((.((((((.....)))))).)).)).)).))))).)))))....(((..(((((.((((....)))).))))).)))........---- ( -23.50) >consensus UUUUCACUUUGACAAUGCCGAC_CGAUUCGAGUCGA_____GUCGACUGCCAAAGUGAAAACUUUGCCUCAAAUUGAAAUGCACAUUUUAAUUUCGGCCCGAAACC____ .((((.(((((.......(.....)......(((((......)))))...))))).)))).....(((..((((((((((....)))))))))).)))............ (-15.23 = -16.30 + 1.07)


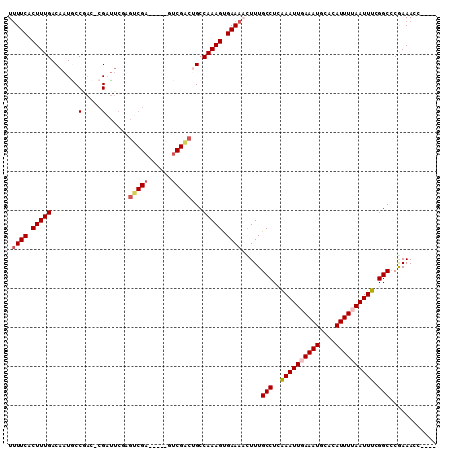
| Location | 13,488,465 – 13,488,565 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.85 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13488465 100 - 22407834 ----GGUUUCGGGCCGAAAUUAAAAUGUGCAUUUCAAUUUAAGGCAAAAGUUUCACUUUGGCUGUCGAC-----UCGACGCGAAUCG-GUCGGCAUUGUCAAAGGGAAAA ----........(((.(((((.((((....)))).)))))..))).....((((.((((((((((((((-----.(((......)))-)))))))..))))))).)))). ( -29.60) >DroPse_CAF1 130508 106 - 1 ----GGUUUCGGGCCGAAAUUAAAAUGUGCAUUUAAAUUUGAGGCAAAGUUUUCGCUUUGGCAGCCGACGCGACUCGACUCGAAUCGGGUUGGUAUUGUCAAAGUGAAAA ----........(((.(((((.((((....)))).)))))..)))....((((((((((((((((((((.(((.(((...))).))).))))))..)))))))))))))) ( -39.00) >DroSec_CAF1 87221 100 - 1 ----GGUUUUGGGCCGAAAUUAAAAUGUGCAUUUCAAUUUAAGGCAAAAGUUUCACUUUGGCUGUCGAC-----UCGACGCGAAUCG-GUCGGCAUUGUCAAAGGGAAAA ----...(((..(((.(((((.((((....)))).)))))..)))..)))((((.((((((((((((((-----.(((......)))-)))))))..))))))).)))). ( -29.70) >DroWil_CAF1 138682 91 - 1 AAACGGUUUUGAGCCGAAAUUAAAAUUUUCAUUUCAAUUCGAGGCAAAGUUUUCACUUUGGCAUU------------------GUCG-GGGGGCAUUGUCAAAGUGAACA ((((.((((((((..(((((.((....)).)))))..))))))))...))))(((((((((((.(------------------(((.-...)))).)))))))))))... ( -29.70) >DroAna_CAF1 151417 100 - 1 ----GGUUUCGGGCCGAAAUUAAAAUGUGCAUUUCAAUUUGAGGCAAAGUUUUCACUUUGGCAGCCGAC-----ACGACUAGAAACG-GUCGGCAUUGUCAAAGGGAAAA ----........(((.(((((.((((....)))).)))))..)))....(((((.((((((((((((((-----.((........))-))))))..))))))))))))). ( -31.90) >DroPer_CAF1 106867 106 - 1 ----GGUUUCGGGCCGAAAUUAAAAUGUGCAUUUAAAUUUGAGGCAAAGUUUUCGCUUUGGCAGCCGACGCGACUCGACUGGAAUCGGGUUGGUAUUGUCAAAGUGAAAA ----........(((.(((((.((((....)))).)))))..)))....((((((((((((((((((((.(((.((.....)).))).))))))..)))))))))))))) ( -38.30) >consensus ____GGUUUCGGGCCGAAAUUAAAAUGUGCAUUUCAAUUUGAGGCAAAGUUUUCACUUUGGCAGCCGAC_____UCGACUCGAAUCG_GUCGGCAUUGUCAAAGGGAAAA ............(((..((((.((((....)))).))))...))).....((((.((((((((((((((.....((.....)).....))))))..)))))))).)))). (-20.85 = -21.85 + 1.00)

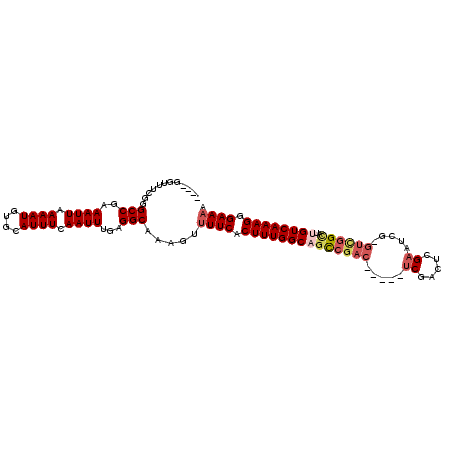
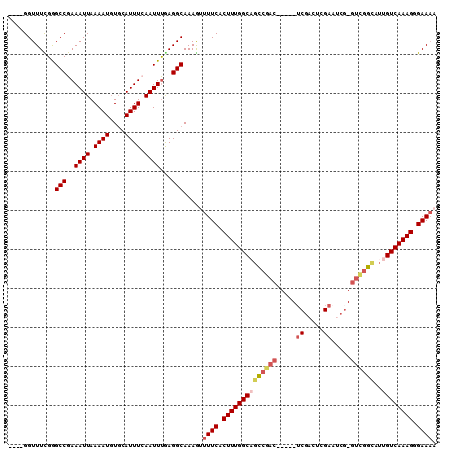
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:57 2006