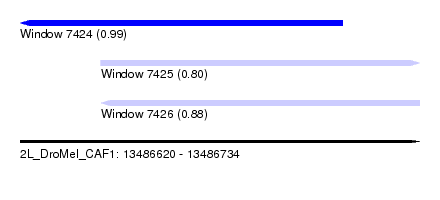
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,486,620 – 13,486,734 |
| Length | 114 |
| Max. P | 0.986961 |

| Location | 13,486,620 – 13,486,712 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -17.08 |
| Consensus MFE | -11.94 |
| Energy contribution | -12.28 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13486620 92 - 22407834 UAG-CAAAAUAGAAUGUACAAUUUCGUUGAUUCGUGGAUCCAGAGAAACUCAACGGAUCCAUGAAAUCAUCUUUCAAAACUAA--ACUAUAAUCA ...-....((((((((........))))(((((((((((((.(((...)))...))))))))).))))...............--.))))..... ( -20.40) >DroSec_CAF1 85386 92 - 1 UAG-CAAAAUAAAAUGUACAAUUUCGUUGAUCCGUGGAUCCGGAAAAACUCAACAGAUCCAUGGAACAAUCUAUCAAAACUAA--UCUAUAAUCA ...-.....................((((.((((((((((.(...........).)))))))))).)))).............--.......... ( -18.40) >DroSim_CAF1 86963 92 - 1 UAG-CAAAGUAAAAUGUACAAUUUCGAUGAUUCGUGGAUCCUGAGAAACUCAACGGAUCCAUGAAACCAUCUUUCAAAACUAA--ACUAUAAUCG ...-....(((.....)))......((((.(((((((((((((((...))))..)))))))))))..))))............--.......... ( -22.50) >DroEre_CAF1 90090 92 - 1 AAG-CAAAAUAAAGUGUAUAAUUUCGUUGAUCAGUGGAUCCUGCAAAACUCAAAAGAUCCACGGAACAAUCUCUCACAACUAA--AGUACAAUCC ...-..........(((((..(((.((((....(((((((...............)))))))(((......)))..)))).))--)))))).... ( -15.56) >DroYak_CAF1 87380 92 - 1 AAG-CAAAUUAAAGUGUAUAAUAUCGUUGAUCCCUGGAUCCUGCAAAAUUCAAUAGAUCCACGGAACAGUCUUUCACAACUAA--AGUCUAAUCC ...-.........(((..........(((.(((.((((((...............)))))).))).))).....)))......--.......... ( -12.32) >DroAna_CAF1 149395 94 - 1 UAGCCAAACUAAUAUCUAAAAUCUGAUUGAUCCAUGGACAUUUGAAGCAAGAAAUAAUCCAUGUACCAGUCGCUAAAAA-UAAGGAUUAAAAUCC ((((.....((.....))......(((((...((((((.((((........))))..))))))...)))))))))....-...((((....)))) ( -13.30) >consensus UAG_CAAAAUAAAAUGUACAAUUUCGUUGAUCCGUGGAUCCUGAAAAACUCAACAGAUCCAUGGAACAAUCUUUCAAAACUAA__ACUAUAAUCC .........................((((.((((((((((...............)))))))))).))))......................... (-11.94 = -12.28 + 0.34)

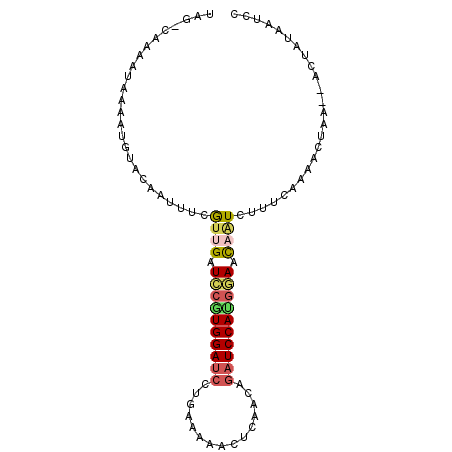
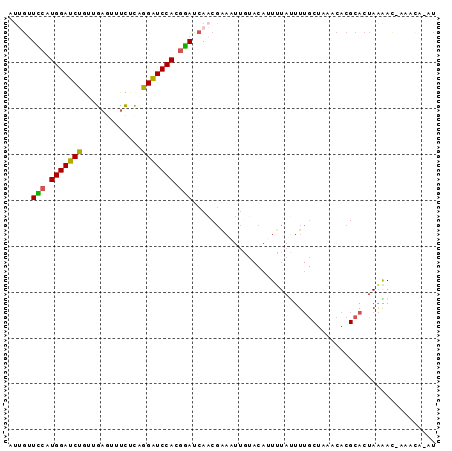
| Location | 13,486,643 – 13,486,734 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 77.02 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -11.98 |
| Energy contribution | -11.32 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13486643 91 + 22407834 AUGAUUUCAUGGAUCCGUUGAGUUUCUCUGGAUCCACGAAUCAACGAAAUUGUACAUUCUAUUUUGCUAAACACGCACUAAGAC-AAACA-AU .((((((..((((((((..(.....)..)))))))).)))))).....(((((....(((....(((.......)))...))).-..)))-)) ( -19.70) >DroPse_CAF1 128518 89 + 1 GUUGAUUCAUGGAUCUGUAGCUGAUCUUAGGGUCCAAGGAUCAACCGAUUAGA--AGUCUAUUU--CCAAACACGCACUAGAACCACACAAAU ((((((((.(((((((..((.....))..))))))).)))))))).((.(((.--...)))..)--).......................... ( -19.00) >DroSec_CAF1 85409 87 + 1 AUUGUUCCAUGGAUCUGUUGAGUUUUUCCGGAUCCACGGAUCAACGAAAUUGUACAUUUUAUUUUGCUAAACACGCACUAAGAC-----A-AU .(((.(((.((((((((..((...))..)))))))).))).)))............(((((...(((.......))).))))).-----.-.. ( -18.10) >DroSim_CAF1 86986 91 + 1 AUGGUUUCAUGGAUCCGUUGAGUUUCUCAGGAUCCACGAAUCAUCGAAAUUGUACAUUUUACUUUGCUAAACACGCACUAAGAC-AAACA-AU (((((((..(((((((..((((...))))))))))).)))))))....(((((...(((((...(((.......))).))))).-..)))-)) ( -20.10) >DroEre_CAF1 90113 91 + 1 AUUGUUCCGUGGAUCUUUUGAGUUUUGCAGGAUCCACUGAUCAACGAAAUUAUACACUUUAUUUUGCUUAACACGCACUAAAAC-AAACA-AU .(((.((.(((((((((...........))))))))).)).)))....................(((.......))).......-.....-.. ( -15.20) >DroYak_CAF1 87403 91 + 1 ACUGUUCCGUGGAUCUAUUGAAUUUUGCAGGAUCCAGGGAUCAACGAUAUUAUACACUUUAAUUUGCUUAACACGCACCAAAAU-AAACA-AC ..((.(((.(((((((.............))))))).))).)).....................(((.......))).......-.....-.. ( -16.52) >consensus AUUGUUCCAUGGAUCUGUUGAGUUUCUCAGGAUCCACGGAUCAACGAAAUUGUACAUUUUAUUUUGCUAAACACGCACUAAAAC_AAACA_AU .....(((.(((((((.............))))))).)))..................................................... (-11.98 = -11.32 + -0.66)

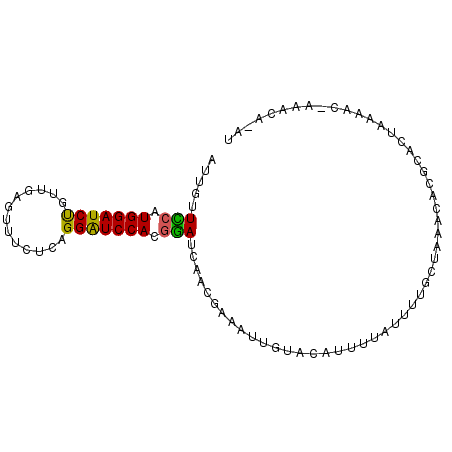
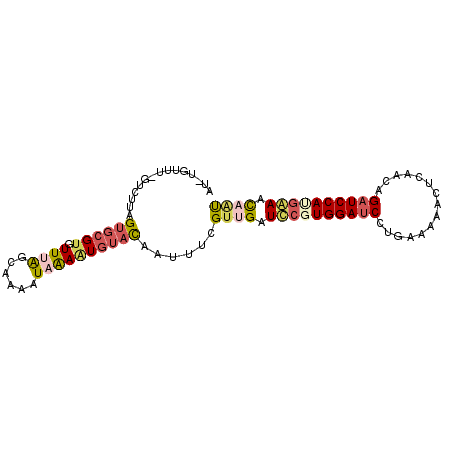
| Location | 13,486,643 – 13,486,734 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 77.02 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -13.01 |
| Energy contribution | -13.79 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13486643 91 - 22407834 AU-UGUUU-GUCUUAGUGCGUGUUUAGCAAAAUAGAAUGUACAAUUUCGUUGAUUCGUGGAUCCAGAGAAACUCAACGGAUCCAUGAAAUCAU ((-(((..-.(((...(((.......)))....)))....))))).....((((((((((((((.(((...)))...))))))))).))))). ( -26.50) >DroPse_CAF1 128518 89 - 1 AUUUGUGUGGUUCUAGUGCGUGUUUGG--AAAUAGACU--UCUAAUCGGUUGAUCCUUGGACCCUAAGAUCAGCUACAGAUCCAUGAAUCAAC ......((((.(((.(((((...((((--((......)--))))).))(((((((.((((...))))))))))))))))).))))........ ( -20.90) >DroSec_CAF1 85409 87 - 1 AU-U-----GUCUUAGUGCGUGUUUAGCAAAAUAAAAUGUACAAUUUCGUUGAUCCGUGGAUCCGGAAAAACUCAACAGAUCCAUGGAACAAU ..-.-----......((((((.((((......))))))))))......((((.((((((((((.(...........).)))))))))).)))) ( -24.60) >DroSim_CAF1 86986 91 - 1 AU-UGUUU-GUCUUAGUGCGUGUUUAGCAAAGUAAAAUGUACAAUUUCGAUGAUUCGUGGAUCCUGAGAAACUCAACGGAUCCAUGAAACCAU ..-.....-......((((((.((((......))))))))))...........(((((((((((((((...))))..)))))))))))..... ( -24.30) >DroEre_CAF1 90113 91 - 1 AU-UGUUU-GUUUUAGUGCGUGUUAAGCAAAAUAAAGUGUAUAAUUUCGUUGAUCAGUGGAUCCUGCAAAACUCAAAAGAUCCACGGAACAAU ((-(((.(-..((((.(((.......)))...))))..).)))))...((((.((.(((((((...............))))))).)).)))) ( -18.16) >DroYak_CAF1 87403 91 - 1 GU-UGUUU-AUUUUGGUGCGUGUUAAGCAAAUUAAAGUGUAUAAUAUCGUUGAUCCCUGGAUCCUGCAAAAUUCAAUAGAUCCACGGAACAGU ((-(((.(-((((((.(((.......)))...))))))).)))))....(((.(((.((((((...............)))))).))).))). ( -19.66) >consensus AU_UGUUU_GUCUUAGUGCGUGUUUAGCAAAAUAAAAUGUACAAUUUCGUUGAUCCGUGGAUCCUGAAAAACUCAACAGAUCCAUGAAACAAU ...............((((((.((((......))))))))))......((((.((((((((((...............)))))))))).)))) (-13.01 = -13.79 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:51 2006