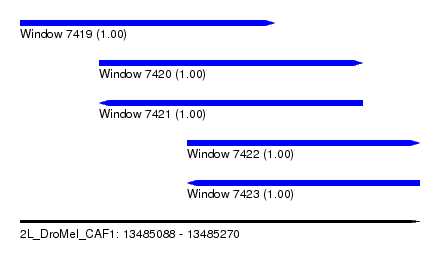
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,485,088 – 13,485,270 |
| Length | 182 |
| Max. P | 0.999985 |

| Location | 13,485,088 – 13,485,204 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.87 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13485088 116 + 22407834 ---AAAAAAACAAC-ACCAAGAUGCAGGCCGCAUUGAUGCUAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCA ---...........-.....(((((.....)))))..(((((...((((....)))).)))))...........((((((...((((((..(..........)..))))))..)))))). ( -27.10) >DroVir_CAF1 134476 117 + 1 ---ACAACAACAACAAACAAGAUGCAGGCCGCAUUGAUGCUAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCA ---.................(((((.....)))))..(((((...((((....)))).)))))...........((((((...((((((..(..........)..))))))..)))))). ( -27.10) >DroGri_CAF1 92985 117 + 1 ---ACAACAACAACAAACAAGAUGCAGGCCGCAUUGAUGCUAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCA ---.................(((((.....)))))..(((((...((((....)))).)))))...........((((((...((((((..(..........)..))))))..)))))). ( -27.10) >DroEre_CAF1 88563 115 + 1 ---AAA-AAACAAC-ACCAAGAUGCAGGCCGCAUUGAUGCUAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCA ---...-.......-.....(((((.....)))))..(((((...((((....)))).)))))...........((((((...((((((..(..........)..))))))..)))))). ( -27.10) >DroWil_CAF1 134224 115 + 1 ---AAC-AAAAAAA-AACAAGAUGCAGGCCGCAUUGAUGCUAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCA ---...-.......-.....(((((.....)))))..(((((...((((....)))).)))))...........((((((...((((((..(..........)..))))))..)))))). ( -27.10) >DroMoj_CAF1 113362 120 + 1 AGAACAACAACAACAAACAAGAUGCAGGCCGCAUUGAUGCUAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCA ....................(((((.....)))))..(((((...((((....)))).)))))...........((((((...((((((..(..........)..))))))..)))))). ( -27.10) >consensus ___AAAAAAACAAC_AACAAGAUGCAGGCCGCAUUGAUGCUAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCA ....................(((((.....)))))..(((((...((((....)))).)))))...........((((((...((((((..(..........)..))))))..)))))). (-27.10 = -27.10 + -0.00)

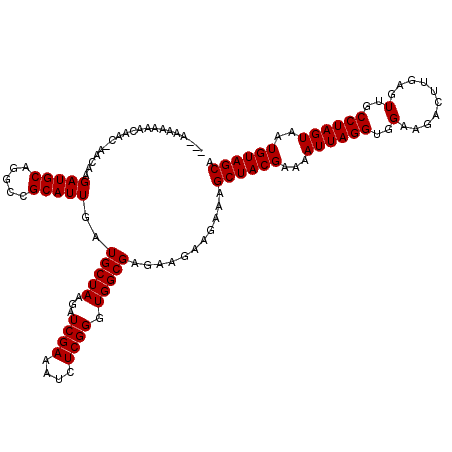
| Location | 13,485,124 – 13,485,244 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.72 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -35.15 |
| Energy contribution | -35.32 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.96 |
| SVM RNA-class probability | 0.999965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13485124 120 + 22407834 UAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGU ............(((((((((((((.........((((((...((((((..(..........)..))))))..)))))).......(((((......)))))))))....))))))))). ( -35.90) >DroEre_CAF1 88598 120 + 1 UAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGU ............(((((((((((((.........((((((...((((((..(..........)..))))))..)))))).......(((((......)))))))))....))))))))). ( -35.90) >DroWil_CAF1 134259 120 + 1 UAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCAGAGU ..((((....)))).((((((((((.........((((((...((((((..(..........)..))))))..)))))).......(((((......)))))))))))))))........ ( -32.20) >DroYak_CAF1 85887 120 + 1 UAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGU ............(((((((((((((.........((((((...((((((..(..........)..))))))..)))))).......(((((......)))))))))....))))))))). ( -35.90) >DroMoj_CAF1 113402 120 + 1 UAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGU ............(((((((((((((.........((((((...((((((..(..........)..))))))..)))))).......(((((......)))))))))....))))))))). ( -35.90) >DroAna_CAF1 147877 120 + 1 UAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGU ............(((((((((((((.........((((((...((((((..(..........)..))))))..)))))).......(((((......)))))))))....))))))))). ( -35.90) >consensus UAAGAUCGAAAUCUCGGGUGGCGAGAAGAAGAAAGCUACGAAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGU ............(((((((((((((.........((((((...((((((..(..........)..))))))..)))))).......(((((......)))))))))....))))))))). (-35.15 = -35.32 + 0.17)

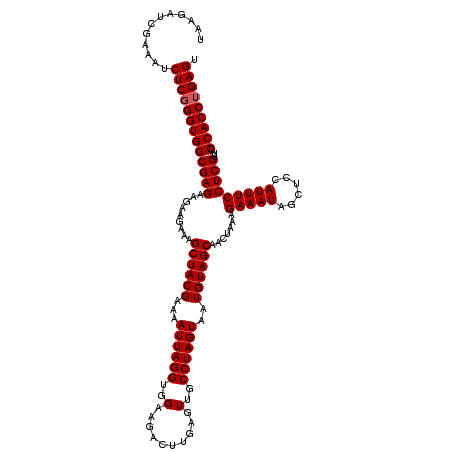
| Location | 13,485,124 – 13,485,244 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.72 |
| Mean single sequence MFE | -33.61 |
| Consensus MFE | -33.61 |
| Energy contribution | -33.61 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.39 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.38 |
| SVM RNA-class probability | 0.999985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13485124 120 - 22407834 ACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUUCGUAGCUUUCUUCUUCUCGCCACCCGAGAUUUCGAUCUUA .(((.(((((....((((((((..(((((((..(((.(((((((.........))))))).))).................)))))))..)).))))))))))).)))............ ( -33.61) >DroEre_CAF1 88598 120 - 1 ACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUUCGUAGCUUUCUUCUUCUCGCCACCCGAGAUUUCGAUCUUA .(((.(((((....((((((((..(((((((..(((.(((((((.........))))))).))).................)))))))..)).))))))))))).)))............ ( -33.61) >DroWil_CAF1 134259 120 - 1 ACUCUGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUUCGUAGCUUUCUUCUUCUCGCCACCCGAGAUUUCGAUCUUA .(((.(((((....((((((((..(((((((..(((.(((((((.........))))))).))).................)))))))..)).))))))))))).)))............ ( -33.61) >DroYak_CAF1 85887 120 - 1 ACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUUCGUAGCUUUCUUCUUCUCGCCACCCGAGAUUUCGAUCUUA .(((.(((((....((((((((..(((((((..(((.(((((((.........))))))).))).................)))))))..)).))))))))))).)))............ ( -33.61) >DroMoj_CAF1 113402 120 - 1 ACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUUCGUAGCUUUCUUCUUCUCGCCACCCGAGAUUUCGAUCUUA .(((.(((((....((((((((..(((((((..(((.(((((((.........))))))).))).................)))))))..)).))))))))))).)))............ ( -33.61) >DroAna_CAF1 147877 120 - 1 ACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUUCGUAGCUUUCUUCUUCUCGCCACCCGAGAUUUCGAUCUUA .(((.(((((....((((((((..(((((((..(((.(((((((.........))))))).))).................)))))))..)).))))))))))).)))............ ( -33.61) >consensus ACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUUCGUAGCUUUCUUCUUCUCGCCACCCGAGAUUUCGAUCUUA .(((.(((((....((((((((..(((((((..(((.(((((((.........))))))).))).................)))))))..)).))))))))))).)))............ (-33.61 = -33.61 + -0.00)

| Location | 13,485,164 – 13,485,270 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -24.39 |
| Energy contribution | -24.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.54 |
| SVM RNA-class probability | 0.999917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13485164 106 + 22407834 AAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGUCAUGGGUAAUAAUAU--AA------AAUACCAAA------ ....(((((((((.(((...((((((.((....))..))))))...(((((......)))))...))).)))))))))......((((.......--..------..))))...------ ( -26.40) >DroSim_CAF1 85500 106 + 1 AAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGUCAUGGGUAAUAAUAU--AA------AAUACCAAA------ ....(((((((((.(((...((((((.((....))..))))))...(((((......)))))...))).)))))))))......((((.......--..------..))))...------ ( -26.40) >DroEre_CAF1 88638 106 + 1 AAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGUCAUGGGUAAUAAUAU--AA------AAUACCAAA------ ....(((((((((.(((...((((((.((....))..))))))...(((((......)))))...))).)))))))))......((((.......--..------..))))...------ ( -26.40) >DroWil_CAF1 134299 120 + 1 AAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCAGAGUCAUGGGUAAUAAUAAUUAACAAAUUAAAACCAAAAAAGAA .......((((((.(((...((((((.((....))..))))))...(((((......)))))...))).)))))).......(((.((((............))))...)))........ ( -23.00) >DroYak_CAF1 85927 106 + 1 AAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGUCAUGGGUAAUAAUAU--AA------AAUACCAAA------ ....(((((((((.(((...((((((.((....))..))))))...(((((......)))))...))).)))))))))......((((.......--..------..))))...------ ( -26.40) >DroAna_CAF1 147917 106 + 1 AAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGUCAUGGGUAAUAAUAU--AA------AAUACCAAA------ ....(((((((((.(((...((((((.((....))..))))))...(((((......)))))...))).)))))))))......((((.......--..------..))))...------ ( -26.40) >consensus AAAAUUAGGUGGAAGACUUGAGUUGCCUAGUAAUGUAGCAACUAAAGAAAUAGCUCCAUUUCCUCGUUAUCCACCUGAGUCAUGGGUAAUAAUAU__AA______AAUACCAAA______ ....(((((((((.(((...((((((...........))))))...(((((......)))))...))).)))))))))......((((...................))))......... (-24.39 = -24.73 + 0.33)

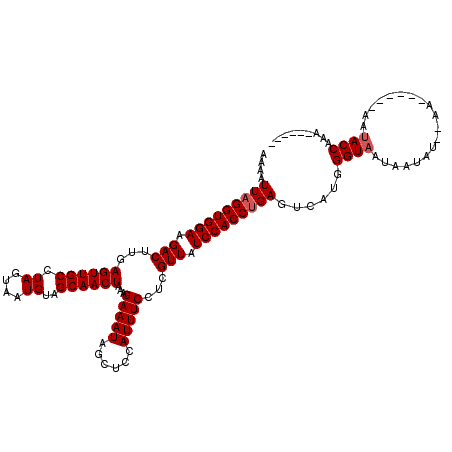
| Location | 13,485,164 – 13,485,270 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -27.46 |
| Energy contribution | -27.79 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.95 |
| SVM RNA-class probability | 0.999964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13485164 106 - 22407834 ------UUUGGUAUU------UU--AUAUUAUUACCCAUGACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUU ------...((((..------..--.......))))........(((((((..(((((((((((....)))))))))(((((((.........)))))))...))..)))))))...... ( -29.30) >DroSim_CAF1 85500 106 - 1 ------UUUGGUAUU------UU--AUAUUAUUACCCAUGACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUU ------...((((..------..--.......))))........(((((((..(((((((((((....)))))))))(((((((.........)))))))...))..)))))))...... ( -29.30) >DroEre_CAF1 88638 106 - 1 ------UUUGGUAUU------UU--AUAUUAUUACCCAUGACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUU ------...((((..------..--.......))))........(((((((..(((((((((((....)))))))))(((((((.........)))))))...))..)))))))...... ( -29.30) >DroWil_CAF1 134299 120 - 1 UUCUUUUUUGGUUUUAAUUUGUUAAUUAUUAUUACCCAUGACUCUGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUU .........(((..((((.........))))..))).........((((((..(((((((((((....)))))))))(((((((.........)))))))...))..))))))....... ( -29.00) >DroYak_CAF1 85927 106 - 1 ------UUUGGUAUU------UU--AUAUUAUUACCCAUGACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUU ------...((((..------..--.......))))........(((((((..(((((((((((....)))))))))(((((((.........)))))))...))..)))))))...... ( -29.30) >DroAna_CAF1 147917 106 - 1 ------UUUGGUAUU------UU--AUAUUAUUACCCAUGACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUU ------...((((..------..--.......))))........(((((((..(((((((((((....)))))))))(((((((.........)))))))...))..)))))))...... ( -29.30) >consensus ______UUUGGUAUU______UU__AUAUUAUUACCCAUGACUCAGGUGGAUAACGAGGAAAUGGAGCUAUUUCUUUAGUUGCUACAUUACUAGGCAACUCAAGUCUUCCACCUAAUUUU .........((((...................))))........(((((((..(((((((((((....)))))))))(((((((.........)))))))...))..)))))))...... (-27.46 = -27.79 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:49 2006