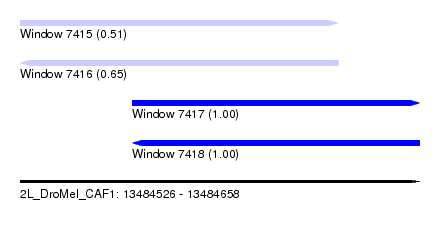
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,484,526 – 13,484,658 |
| Length | 132 |
| Max. P | 0.999405 |

| Location | 13,484,526 – 13,484,631 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13484526 105 + 22407834 AGAAAAG--CAGCCGACCACUGGCGAU-CAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUUCGAGGCUGACGCAGUUUUUCAAACGCCAC .((((((--(.((.((.....(((...-.......))).....)).))..))......((((((.(((((..(((....)))))))).)))))))))))......... ( -30.80) >DroVir_CAF1 133979 97 + 1 AGA-AAGUAGAGCCGACCACUUGCGAUUCAACGCAGCACAUCCUCCGCCCGCCACUCAACUGCGCCAGGA------CCACAACACUGACGCAGUUCCUCAAC----AC ...-..((.(((........(((((......))))).....................(((((((.(((..------........))).))))))).))).))----.. ( -20.60) >DroPse_CAF1 126248 94 + 1 AGAUACG--CAGCCGACCACUGGCGAU-CAACGCAGCCCAUCCUCAGCCCGCCACUCAGCUGCGCCAGCCC--------CCGCGCUGACGCAGUUUUUCAA---CCAC .((...(--(.((.((.....(((...-.......))).....)).))..))...))(((((((.((((..--------....)))).)))))))......---.... ( -23.60) >DroGri_CAF1 92463 96 + 1 AGA-AAGCACAGCCGACCACUGGCAAU-CAACGCAGCACAUCCUCAGCCCGCCACUCAGCUGCGCCAGGA------CCUAAUCACUGACGCAGUUGCUCAAC----AC .((-..((...((((.....))))...-.......((.........))..))....((((((((.(((((------.....)).))).)))))))).))...----.. ( -22.20) >DroAna_CAF1 147290 98 + 1 AGAUAAG--CAGCCGACCACUGGCGAU-CAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAGUC-----ACUUC-CGGCUGACGCAGUUUUCCAACAACCA- .((...(--(.((.((.....(((...-.......))).....)).))..))...))(((((((.(((((-----.....-.))))).)))))))............- ( -24.80) >DroPer_CAF1 100813 94 + 1 AGAUACG--CAGCCGACCACUAGCGAU-CAACGCAGCCCAUCCUCAGCCCGCCACUCAGCUGCGCCAGCCC--------CCGCGCUGACGCAGUUUUUCAA---CCAC .(((.((--(((.......)).)))))-)...((.((.........))..)).....(((((((.((((..--------....)))).)))))))......---.... ( -21.90) >consensus AGAUAAG__CAGCCGACCACUGGCGAU_CAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAGCC______C_UCAACGCUGACGCAGUUUUUCAAC__CCAC ...........((((.....))))........((.((.........))..)).....(((((((.((((..............)))).)))))))............. (-17.19 = -17.52 + 0.33)


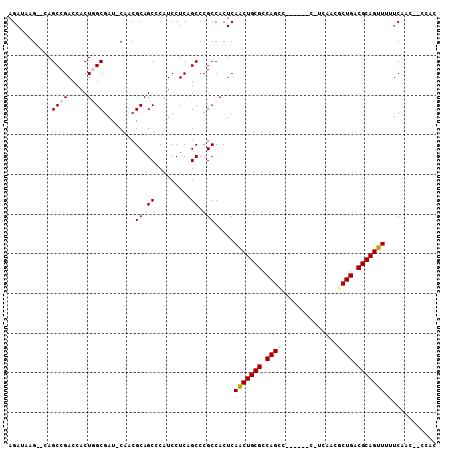
| Location | 13,484,526 – 13,484,631 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -39.33 |
| Consensus MFE | -25.69 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13484526 105 - 22407834 GUGGCGUUUGAAAAACUGCGUCAGCCUCGAAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGAUGGGCUGCGUUG-AUCGCCAGUGGUCGGCUG--CUUUUCU .(((((.(..(..((((((((((((((((....)))..))))))))))))).....((((.((......)).)))).)..-).)))))(..(....)..--)...... ( -46.50) >DroVir_CAF1 133979 97 - 1 GU----GUUGAGGAACUGCGUCAGUGUUGUGG------UCCUGGCGCAGUUGAGUGGCGGGCGGAGGAUGUGCUGCGUUGAAUCGCAAGUGGUCGGCUCUACUU-UCU ..----.......(((((((((((..(....)------..)))))))))))((((((...((((........))))(((((..(....)...))))).))))))-... ( -32.80) >DroPse_CAF1 126248 94 - 1 GUGG---UUGAAAAACUGCGUCAGCGCGG--------GGGCUGGCGCAGCUGAGUGGCGGGCUGAGGAUGGGCUGCGUUG-AUCGCCAGUGGUCGGCUG--CGUAUCU (..(---((....)))..)((((((....--------..))))))((((((((.(.((.(((.((.((((.....)))).-.))))).)).))))))))--)...... ( -40.60) >DroGri_CAF1 92463 96 - 1 GU----GUUGAGCAACUGCGUCAGUGAUUAGG------UCCUGGCGCAGCUGAGUGGCGGGCUGAGGAUGUGCUGCGUUG-AUUGCCAGUGGUCGGCUGUGCUU-UCU ..----...(((((...(((((((.(((...)------)))))))))((((((.(.((.(((....((((.....)))).-...))).)).))))))).)))))-... ( -34.50) >DroAna_CAF1 147290 98 - 1 -UGGUUGUUGGAAAACUGCGUCAGCCG-GAAGU-----GACUGGCGCAGUUGAGUGGCGGGCUGAGGAUGGGCUGCGUUG-AUCGCCAGUGGUCGGCUG--CUUAUCU -.(.((((((...(((((((((((((.-...).-----).)))))))))))...)))))).)...((((((((...((((-((((....)))))))).)--))))))) ( -41.90) >DroPer_CAF1 100813 94 - 1 GUGG---UUGAAAAACUGCGUCAGCGCGG--------GGGCUGGCGCAGCUGAGUGGCGGGCUGAGGAUGGGCUGCGUUG-AUCGCUAGUGGUCGGCUG--CGUAUCU (..(---((....)))..)((((((....--------..))))))((((((.....((((.((......)).))))...(-((((....))))))))))--)...... ( -39.70) >consensus GUGG__GUUGAAAAACUGCGUCAGCGCGGA_G______GGCUGGCGCAGCUGAGUGGCGGGCUGAGGAUGGGCUGCGUUG_AUCGCCAGUGGUCGGCUG__CUUAUCU ...............(((((((((................)))))))))......(((.((((........((((((......))).))))))).))).......... (-25.69 = -25.72 + 0.03)

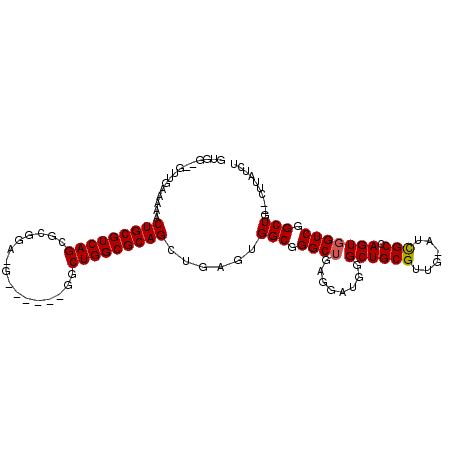

| Location | 13,484,563 – 13,484,658 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13484563 95 + 22407834 UCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUUCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCCCAAAAAACC .....(((..((.....(((((((.(((((..(((....)))))))).)))))))........))....)))....................... ( -23.02) >DroSec_CAF1 83336 94 + 1 UCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUUCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCC-CCAAAAAACC .....(((..((.....(((((((.(((((..(((....)))))))).)))))))........))....)))............-.......... ( -23.02) >DroSim_CAF1 84899 95 + 1 UCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUUCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCCCAAAAAACC .....(((..((.....(((((((.(((((..(((....)))))))).)))))))........))....)))....................... ( -23.02) >DroEre_CAF1 88040 95 + 1 UCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGAUUCCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCCCAAAAAACC .....(((..((.....(((((((.(((((..(((....)))))))).)))))))........))....)))....................... ( -22.42) >DroYak_CAF1 85304 95 + 1 UCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUCCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCCCAAAAAACC .....(((..((.....(((((((.(((((..(((....)))))))).)))))))........))....)))....................... ( -22.42) >DroAna_CAF1 147327 85 + 1 UCCUCAGCCCGCCACUCAACUGCGCCAGUC-----ACUUC-CGGCUGACGCAGUUUUCCAACAACCA-GGCUACAGAAAGAA---AAAAAAAACC .....((((........(((((((.(((((-----.....-.))))).)))))))............-))))..........---.......... ( -20.35) >consensus UCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUUCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCCCAAAAAACC .....(((..((.....(((((((.(((((..(((....)))))))).)))))))........))....)))....................... (-20.10 = -20.50 + 0.40)


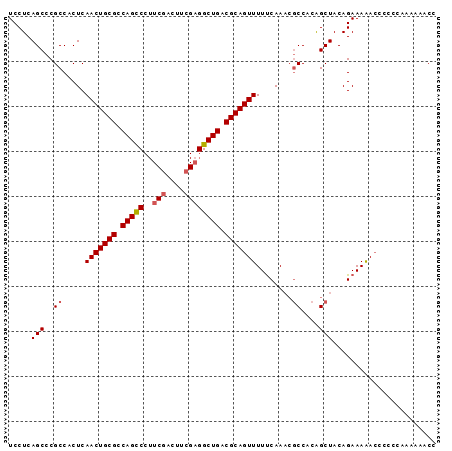
| Location | 13,484,563 – 13,484,658 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -32.03 |
| Energy contribution | -32.63 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13484563 95 - 22407834 GGUUUUUUGGGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGAAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGA ..................(((.(((((((.((........((((((((((((((((....)))..)))))))))))))..)).)).))))).))) ( -36.10) >DroSec_CAF1 83336 94 - 1 GGUUUUUUGG-GGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGAAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGA ..........-.......(((.(((((((.((........((((((((((((((((....)))..)))))))))))))..)).)).))))).))) ( -36.10) >DroSim_CAF1 84899 95 - 1 GGUUUUUUGGGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGAAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGA ..................(((.(((((((.((........((((((((((((((((....)))..)))))))))))))..)).)).))))).))) ( -36.10) >DroEre_CAF1 88040 95 - 1 GGUUUUUUGGGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGGAAUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGA ..................(((.(((((((.((........((((((((((((((((....)))..)))))))))))))..)).)).))))).))) ( -35.20) >DroYak_CAF1 85304 95 - 1 GGUUUUUUGGGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGGAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGA ..................(((.(((((((.((........((((((((((((((((....)))..)))))))))))))..)).)).))))).))) ( -35.20) >DroAna_CAF1 147327 85 - 1 GGUUUUUUUU---UUCUUUCUGUAGCC-UGGUUGUUGGAAAACUGCGUCAGCCG-GAAGU-----GACUGGCGCAGUUGAGUGGCGGGCUGAGGA ..........---.....(((.(((((-((.(..((....(((((((((((((.-...).-----).))))))))))).))..)))))))).))) ( -34.00) >consensus GGUUUUUUGGGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGAAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGA ..................(((.(((((....((((.....((((((((((((((((....)))..)))))))))))))....))))))))).))) (-32.03 = -32.63 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:44 2006