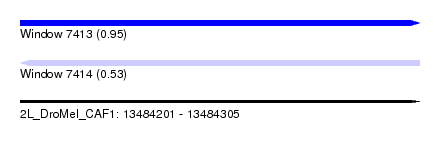
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,484,201 – 13,484,305 |
| Length | 104 |
| Max. P | 0.954076 |

| Location | 13,484,201 – 13,484,305 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -18.77 |
| Energy contribution | -20.38 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
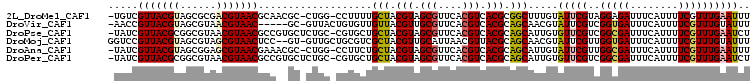
>2L_DroMel_CAF1 13484201 104 + 22407834 AAAUUCAAACGAAAAUGAAAUCUCCUACGAAUACAAAGCCGCGUGACGUGAACGCUACGUAGCAAAAGG-CCAG-GCGUUGCGUUACGUCGCGCUACGUAACGACA- ........(((.........((......))..........(((((((((((.(((.((((.((.....)-)...-)))).))))))))))))))..))).......- ( -30.90) >DroVir_CAF1 133678 100 + 1 AAAUACAAACGAAAAUGAAAUCACCGACGAAUACGUUGCUGCGUGACGUGAACGCAACGUAACAACACAGUAAC-GC-----GUUACGUUACGCUACGUAACGGUU- .................................((((((.(((((((((((.(((...((.((......)).))-))-----))))))))))))...))))))...- ( -28.00) >DroPse_CAF1 125808 105 + 1 AGAUUCAAACGAAAAUGAAAUCGCCGACGAACACAAUGCUGCGUGACGUGAACGCUACGUAGCAGCACG-GCAGAGCACGGCGUUACGUUACGCCGCGUAACGAUA- ...((((........))))(((((((..(....)..(((((((((.((....)).)))))))))...))-))...((.((((((......)))))).))...))).- ( -36.60) >DroMoj_CAF1 112434 104 + 1 AAAUACAAACGAAAAUGAAAUCACCAACGAAUACGUUGCUGCGUAACGUUAAUGCAACGUAGCGACGCAGCAAC-AC--GGAGUUACGCUACGCUACGUAACGGACC .......................((.........((((((((((.(((((.....)))))....))))))))))-..--...((((((........))))))))... ( -30.30) >DroAna_CAF1 146866 104 + 1 AAAUUCAAACGAAAAUGAAAUCGCCAACGAAUACAAUGCUGCGUGACGUGAACGCUACGUAGCAGAAGG-CCAG-GCGUUUCGUUACGCUCCGCUACGUAACGAUA- ...((((........))))((((...(((.......(((((((((.((....)).)))))))))...((-(..(-((((......)))))..))).)))..)))).- ( -31.80) >DroPer_CAF1 100376 105 + 1 AGAUUCAAACGAAAAUGAAAUCGCCGACGAACACAAUGCUGCGUGACGUGAACGCUACGUAGCAGCACG-GCAGAGCACGGCGUUACGUUACGCCGCGUAACGAUA- ...((((........))))(((((((..(....)..(((((((((.((....)).)))))))))...))-))...((.((((((......)))))).))...))).- ( -36.60) >consensus AAAUUCAAACGAAAAUGAAAUCGCCGACGAAUACAAUGCUGCGUGACGUGAACGCUACGUAGCAACACG_CCAG_GC__GGCGUUACGUUACGCUACGUAACGAUA_ ...................(((..............((((((((..((....))..))))))))..................((((((........))))))))).. (-18.77 = -20.38 + 1.61)


| Location | 13,484,201 – 13,484,305 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13484201 104 - 22407834 -UGUCGUUACGUAGCGCGACGUAACGCAACGC-CUGG-CCUUUUGCUACGUAGCGUUCACGUCACGCGGCUUUGUAUUCGUAGGAGAUUUCAUUUUCGUUUGAAUUU -.........((.(((.((((((((((.(((.-.(((-(.....))))))).))))).))))).))).)).....(((((.(.((((......)))).).))))).. ( -31.50) >DroVir_CAF1 133678 100 - 1 -AACCGUUACGUAGCGUAACGUAAC-----GC-GUUACUGUGUUGUUACGUUGCGUUCACGUCACGCAGCAACGUAUUCGUCGGUGAUUUCAUUUUCGUUUGUAUUU -...(((.(((((((((((((((((-----((-(......)).)))))))))))))).)))).)))((.(.(((....))).).))..................... ( -29.30) >DroPse_CAF1 125808 105 - 1 -UAUCGUUACGCGGCGUAACGUAACGCCGUGCUCUGC-CGUGCUGCUACGUAGCGUUCACGUCACGCAGCAUUGUGUUCGUCGGCGAUUUCAUUUUCGUUUGAAUCU -.((((((.((((((((......))))))))....((-.(((((((...((..((....))..))))))))).)).......))))))((((........))))... ( -34.40) >DroMoj_CAF1 112434 104 - 1 GGUCCGUUACGUAGCGUAGCGUAACUCC--GU-GUUGCUGCGUCGCUACGUUGCAUUAACGUUACGCAGCAACGUAUUCGUUGGUGAUUUCAUUUUCGUUUGUAUUU ((...(((((((......))))))).))--..-((((((((((....((((((...)))))).))))))))))((((.((..((((....))))..))...)))).. ( -35.20) >DroAna_CAF1 146866 104 - 1 -UAUCGUUACGUAGCGGAGCGUAACGAAACGC-CUGG-CCUUCUGCUACGUAGCGUUCACGUCACGCAGCAUUGUAUUCGUUGGCGAUUUCAUUUUCGUUUGAAUUU -...((((((((((((((((((......))))-....-...))))))))))))))....(((((((.(........).)).)))))..((((........))))... ( -31.51) >DroPer_CAF1 100376 105 - 1 -UAUCGUUACGCGGCGUAACGUAACGCCGUGCUCUGC-CGUGCUGCUACGUAGCGUUCACGUCACGCAGCAUUGUGUUCGUCGGCGAUUUCAUUUUCGUUUGAAUCU -.((((((.((((((((......))))))))....((-.(((((((...((..((....))..))))))))).)).......))))))((((........))))... ( -34.40) >consensus _UAUCGUUACGUAGCGUAACGUAACGCC__GC_CUGG_CGUGCUGCUACGUAGCGUUCACGUCACGCAGCAUUGUAUUCGUCGGCGAUUUCAUUUUCGUUUGAAUUU .....(((((((......)))))))...................(((.(((.(((....))).))).))).....(((((..(((((........)))))))))).. (-17.66 = -17.88 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:41 2006