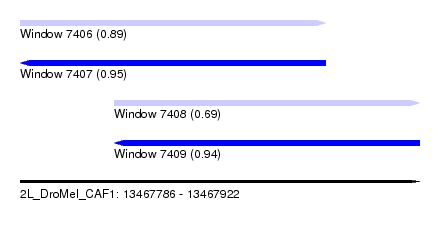
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,467,786 – 13,467,922 |
| Length | 136 |
| Max. P | 0.947442 |

| Location | 13,467,786 – 13,467,890 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 99.36 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -39.20 |
| Energy contribution | -42.53 |
| Covariance contribution | 3.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13467786 104 + 22407834 UGCUCUUUCGAUUUCCACCUGGCCGGAUGGCUGAUUCAGCGACUGGGUGACUGGUUGACUGGCUGACCUGCCAGAGGCAGAGCCACCUGUCAUUGUCAUGGAAU ............(((((.(((((.((.((((.....(((((((..(....)..)))).))))))).)).))))).(((((......))))).......))))). ( -38.90) >DroSec_CAF1 67292 104 + 1 UGCUCUUUCGAUUUCCACCUGGCCGGAUGGCUGAUUCAGCGACUGGGUGACUGGUUGACUGGCUGUCCUGCCAGAGGCAGAGCCACCUGUCAUUGUCAUGGAAU ............(((((.(((((.(((((((.....(((((((..(....)..)))).)))))))))).))))).(((((......))))).......))))). ( -43.80) >DroSim_CAF1 68512 104 + 1 UGCUCUUUCGAUUUCCACCUGGCCGGAUGGCUGAUUCAGCGACUGGGUGACUGGUUGACUGGCUGUCCUGCCAGAGGCAGAGCCACCUGUCAUUGUCAUGGAAU ............(((((.(((((.(((((((.....(((((((..(....)..)))).)))))))))).))))).(((((......))))).......))))). ( -43.80) >consensus UGCUCUUUCGAUUUCCACCUGGCCGGAUGGCUGAUUCAGCGACUGGGUGACUGGUUGACUGGCUGUCCUGCCAGAGGCAGAGCCACCUGUCAUUGUCAUGGAAU ............(((((.(((((.(((((((.....(((((((..(....)..)))).)))))))))).))))).(((((......))))).......))))). (-39.20 = -42.53 + 3.33)

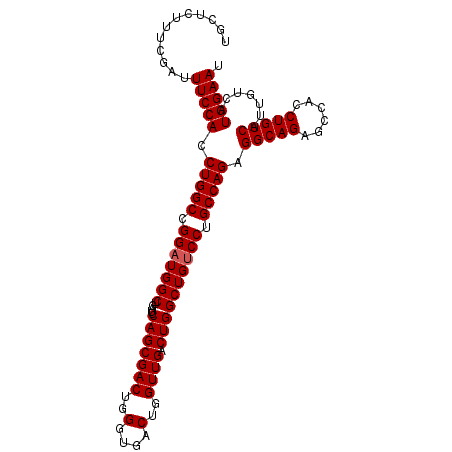
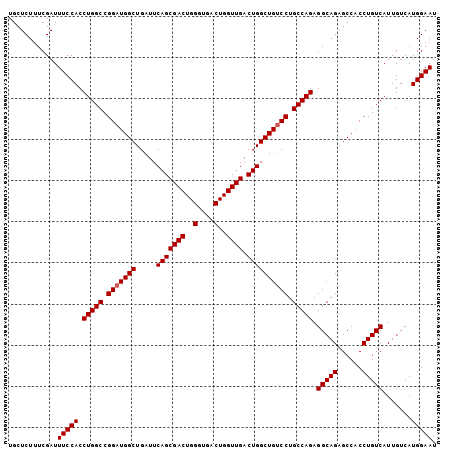
| Location | 13,467,786 – 13,467,890 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 99.36 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -36.80 |
| Energy contribution | -36.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13467786 104 - 22407834 AUUCCAUGACAAUGACAGGUGGCUCUGCCUCUGGCAGGUCAGCCAGUCAACCAGUCACCCAGUCGCUGAAUCAGCCAUCCGGCCAGGUGGAAAUCGAAAGAGCA .((((((..((.((((.(((((((......(((((......)))))......)))))))..)))).)).....(((....)))...)))))).((....))... ( -36.80) >DroSec_CAF1 67292 104 - 1 AUUCCAUGACAAUGACAGGUGGCUCUGCCUCUGGCAGGACAGCCAGUCAACCAGUCACCCAGUCGCUGAAUCAGCCAUCCGGCCAGGUGGAAAUCGAAAGAGCA .((((((..((.((((.(((((((......(((((......)))))......)))))))..)))).)).....(((....)))...)))))).((....))... ( -36.80) >DroSim_CAF1 68512 104 - 1 AUUCCAUGACAAUGACAGGUGGCUCUGCCUCUGGCAGGACAGCCAGUCAACCAGUCACCCAGUCGCUGAAUCAGCCAUCCGGCCAGGUGGAAAUCGAAAGAGCA .((((((..((.((((.(((((((......(((((......)))))......)))))))..)))).)).....(((....)))...)))))).((....))... ( -36.80) >consensus AUUCCAUGACAAUGACAGGUGGCUCUGCCUCUGGCAGGACAGCCAGUCAACCAGUCACCCAGUCGCUGAAUCAGCCAUCCGGCCAGGUGGAAAUCGAAAGAGCA .((((((..((.((((.(((((((......(((((......)))))......)))))))..)))).)).....(((....)))...)))))).((....))... (-36.80 = -36.80 + 0.00)

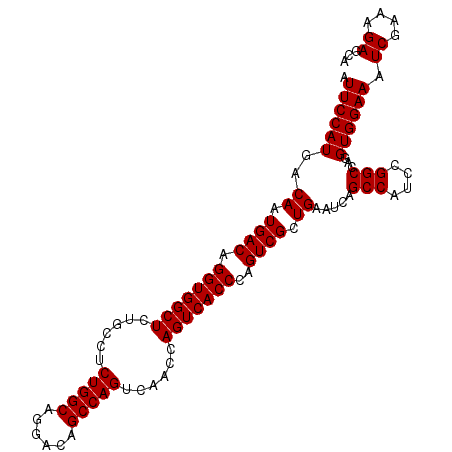
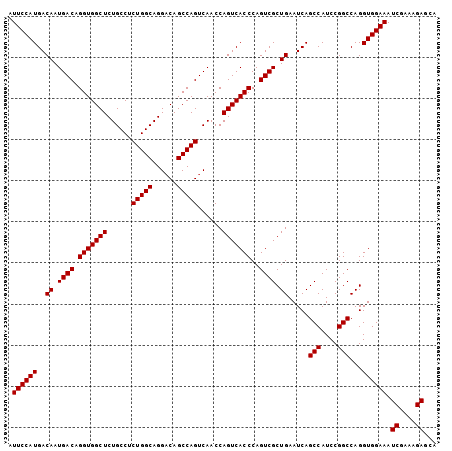
| Location | 13,467,818 – 13,467,922 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -30.49 |
| Energy contribution | -33.50 |
| Covariance contribution | 3.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13467818 104 + 22407834 GAUUCAGCGACUGGGUGACUGGUUGACUGGCUGACCUGCCAGAGGCAGAGCCACCUGUCAUUGUCAUGGAAUGGUAAUGGGAAGGAGAAGGGAAUCGGAAUGGG (((((((((((..(....)..)))).))((((...(((((...))))))))).(((.(((((..(((...)))..)))))..)))......)))))........ ( -37.80) >DroSec_CAF1 67324 104 + 1 GAUUCAGCGACUGGGUGACUGGUUGACUGGCUGUCCUGCCAGAGGCAGAGCCACCUGUCAUUGUCAUGGAAUGGGAAUGGGAAUGGGAAGGCAAUUGGAAUGGG .((((((((((..(....)..)))).))((((...(((((...))))))))).(((.(((((.((((.........)))).)))))..)))......))))... ( -34.50) >DroSim_CAF1 68544 104 + 1 GAUUCAGCGACUGGGUGACUGGUUGACUGGCUGUCCUGCCAGAGGCAGAGCCACCUGUCAUUGUCAUGGAAUGGGAAUGGGAAUGGUAGGGGAAUUGGAAUGGG (((((((((((..(....)..)))).))((((...(((((...))))))))).(((..((((.((((.........)))).))))..))).)))))........ ( -39.30) >consensus GAUUCAGCGACUGGGUGACUGGUUGACUGGCUGUCCUGCCAGAGGCAGAGCCACCUGUCAUUGUCAUGGAAUGGGAAUGGGAAUGGGAAGGGAAUUGGAAUGGG (((((((((((..(....)..)))).))((((...(((((...))))))))).(((.(((((.((((.........)))).)))))..))))))))........ (-30.49 = -33.50 + 3.01)

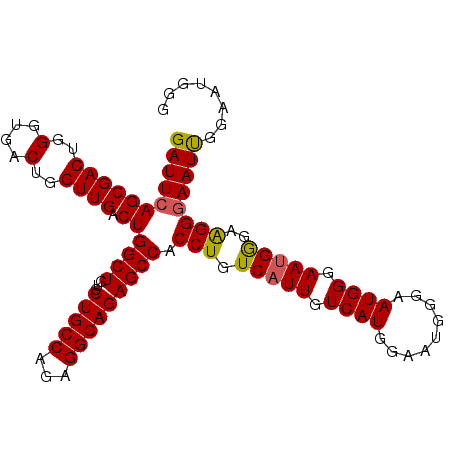
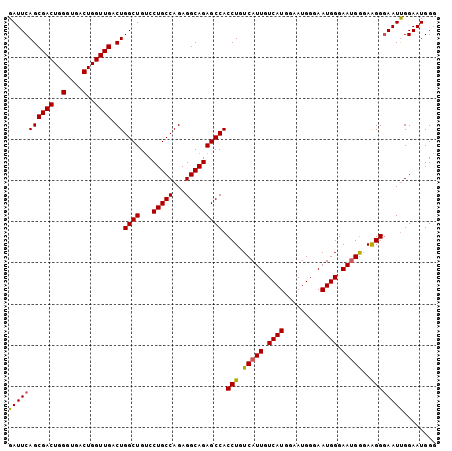
| Location | 13,467,818 – 13,467,922 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -23.90 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13467818 104 - 22407834 CCCAUUCCGAUUCCCUUCUCCUUCCCAUUACCAUUCCAUGACAAUGACAGGUGGCUCUGCCUCUGGCAGGUCAGCCAGUCAACCAGUCACCCAGUCGCUGAAUC .........................................((.((((.(((((((......(((((......)))))......)))))))..)))).)).... ( -23.90) >DroSec_CAF1 67324 104 - 1 CCCAUUCCAAUUGCCUUCCCAUUCCCAUUCCCAUUCCAUGACAAUGACAGGUGGCUCUGCCUCUGGCAGGACAGCCAGUCAACCAGUCACCCAGUCGCUGAAUC .........................................((.((((.(((((((......(((((......)))))......)))))))..)))).)).... ( -23.90) >DroSim_CAF1 68544 104 - 1 CCCAUUCCAAUUCCCCUACCAUUCCCAUUCCCAUUCCAUGACAAUGACAGGUGGCUCUGCCUCUGGCAGGACAGCCAGUCAACCAGUCACCCAGUCGCUGAAUC .........................................((.((((.(((((((......(((((......)))))......)))))))..)))).)).... ( -23.90) >consensus CCCAUUCCAAUUCCCUUCCCAUUCCCAUUCCCAUUCCAUGACAAUGACAGGUGGCUCUGCCUCUGGCAGGACAGCCAGUCAACCAGUCACCCAGUCGCUGAAUC .........................................((.((((.(((((((......(((((......)))))......)))))))..)))).)).... (-23.90 = -23.90 + 0.00)

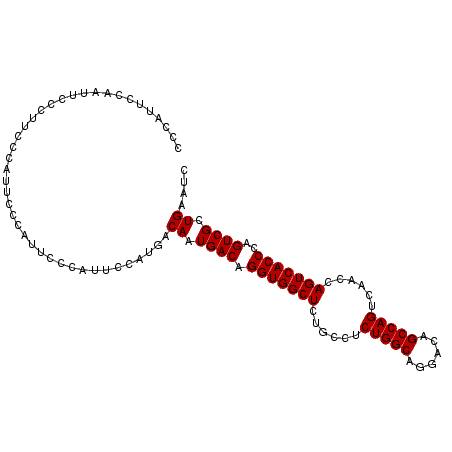
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:36 2006