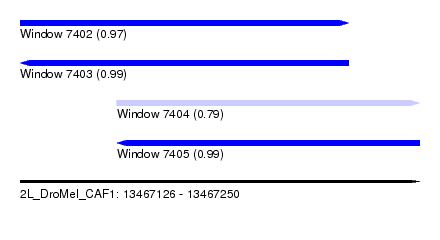
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,467,126 – 13,467,250 |
| Length | 124 |
| Max. P | 0.990113 |

| Location | 13,467,126 – 13,467,228 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -37.56 |
| Consensus MFE | -26.78 |
| Energy contribution | -26.78 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13467126 102 + 22407834 GAUUUCGCGUUCGUGUUUCGGAUUUC-------AGUUUACGGAUUC------CGAAUUCCGGACAUAUACCUGUGUCCGUAUCCGUCUGUCCGUCCGUUCGGACUGCAUAAUUUA ......(((((((.....(((((..(-------((...((((((..------.......((((((((....)))))))).)))))))))...)))))..))))).))........ ( -32.20) >DroSec_CAF1 66632 102 + 1 GAUUUCGCGUUCGUGUUUCGGAUUUC-------GGUUUACGGAUUC------CGAAUUCCGGACAUAUACCUGUGUCCGUAUCCGUCUGUCCGUCCGUUCGGACUGCAUAAUUUA ......(((((((.....(((((..(-------((..((((((...------.....)))(((((((....)))))))))).)))...)))))......))))).))........ ( -34.50) >DroSim_CAF1 67852 102 + 1 GAUUUCGCGUUCGUGUUUCGGAUUUC-------GGUUUACGGAUUC------CGAAUUCCGGACAUAUACCUGUGUCCGUAUCCGUCUGUCCGUCCGUUCGGACUGCAUAAUUUA ......(((((((.....(((((..(-------((..((((((...------.....)))(((((((....)))))))))).)))...)))))......))))).))........ ( -34.50) >DroEre_CAF1 71179 109 + 1 GAUUCCGCGUUCGUGUUUCGGAUUUCGGAUUACGGAUUCCGGAUUC------CGGAAUCCGGACAUAUACCUGUGUCCGCAUCCGUUUGUCCGUCCGCUCGGACUGCAUAAUUUA ......(((((((.((..(((((..(((((...(((((((((...)------))))))))(((((((....)))))))..)))))...)))))...)).))))).))........ ( -48.80) >DroYak_CAF1 67874 115 + 1 GAUUCCGCGUUCGUGUUUCGUGUUUCGGAUUUCGGUUUACGGAUUCGUUUUACGGAUUCCGGACAUAUACCUGUGUCCGUAUCCGUUUGUCCGUCCGUUCGGACUGCAUAAUUUA ((.((((...(((...((((.....))))...)))....)))).)).....((((((..((((((((....)))))))).))))))..(((((......)))))........... ( -37.80) >consensus GAUUUCGCGUUCGUGUUUCGGAUUUC_______GGUUUACGGAUUC______CGAAUUCCGGACAUAUACCUGUGUCCGUAUCCGUCUGUCCGUCCGUUCGGACUGCAUAAUUUA ((...(((....)))..))((((.................((((((.......))))))((((((((....)))))))).))))((..(((((......))))).))........ (-26.78 = -26.78 + -0.00)


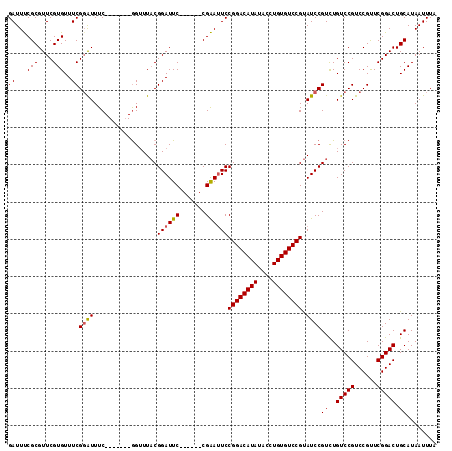
| Location | 13,467,126 – 13,467,228 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -24.88 |
| Energy contribution | -25.04 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13467126 102 - 22407834 UAAAUUAUGCAGUCCGAACGGACGGACAGACGGAUACGGACACAGGUAUAUGUCCGGAAUUCG------GAAUCCGUAAACU-------GAAAUCCGAAACACGAACGCGAAAUC .......(((.(((((......)))))...(((((.((((((........))))))...((((------(..........))-------))))))))..........)))..... ( -28.10) >DroSec_CAF1 66632 102 - 1 UAAAUUAUGCAGUCCGAACGGACGGACAGACGGAUACGGACACAGGUAUAUGUCCGGAAUUCG------GAAUCCGUAAACC-------GAAAUCCGAAACACGAACGCGAAAUC .......(((.(((((......)))))...(((((.((((((........))))))...((((------(..........))-------))))))))..........)))..... ( -30.30) >DroSim_CAF1 67852 102 - 1 UAAAUUAUGCAGUCCGAACGGACGGACAGACGGAUACGGACACAGGUAUAUGUCCGGAAUUCG------GAAUCCGUAAACC-------GAAAUCCGAAACACGAACGCGAAAUC .......(((.(((((......)))))...(((((.((((((........))))))...((((------(..........))-------))))))))..........)))..... ( -30.30) >DroEre_CAF1 71179 109 - 1 UAAAUUAUGCAGUCCGAGCGGACGGACAAACGGAUGCGGACACAGGUAUAUGUCCGGAUUCCG------GAAUCCGGAAUCCGUAAUCCGAAAUCCGAAACACGAACGCGGAAUC .......(((.((((....))))(((....((((((((((((........)))))((((((((------(...))))))))))).)))))...)))...........)))..... ( -41.50) >DroYak_CAF1 67874 115 - 1 UAAAUUAUGCAGUCCGAACGGACGGACAAACGGAUACGGACACAGGUAUAUGUCCGGAAUCCGUAAAACGAAUCCGUAAACCGAAAUCCGAAACACGAAACACGAACGCGGAAUC ...........(((((......)))))..((((((.((((((........))))))..)))))).....((.(((((....((.....)).....((.....))...))))).)) ( -31.60) >consensus UAAAUUAUGCAGUCCGAACGGACGGACAGACGGAUACGGACACAGGUAUAUGUCCGGAAUUCG______GAAUCCGUAAACC_______GAAAUCCGAAACACGAACGCGAAAUC .......(((.(((((......)))))...(((((..(((((........)))))(((.((........)).))).................)))))..........)))..... (-24.88 = -25.04 + 0.16)


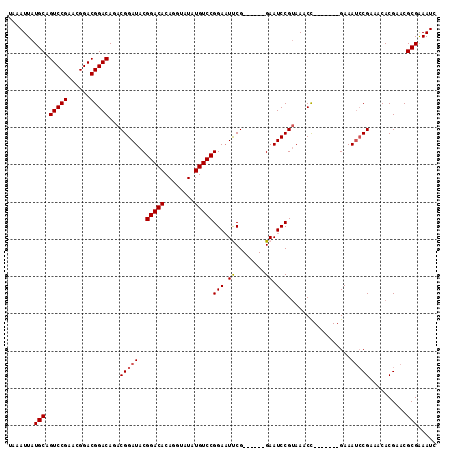
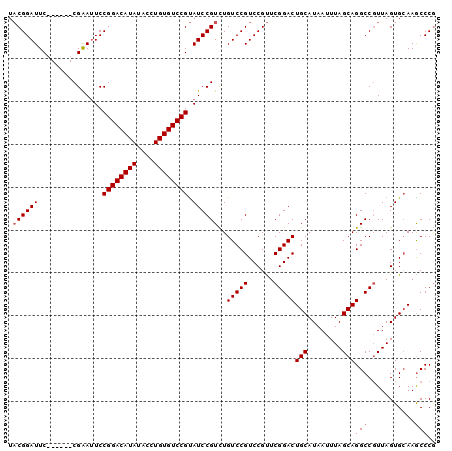
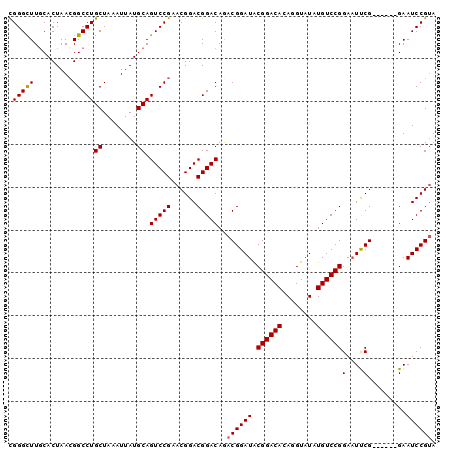
| Location | 13,467,156 – 13,467,250 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 92.95 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.67 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13467156 94 + 22407834 UACGGAUUC------CGAAUUCCGGACAUAUACCUGUGUCCGUAUCCGUCUGUCCGUCCGUUCGGACUGCAUAAUUUAGCAGGCCGUUAGUGCAAACCCG ...((((..------((.((..((((((((....)))))))).)).))...))))(..(...(((.((((........)))).)))...)..)....... ( -28.70) >DroSec_CAF1 66662 94 + 1 UACGGAUUC------CGAAUUCCGGACAUAUACCUGUGUCCGUAUCCGUCUGUCCGUCCGUUCGGACUGCAUAAUUUAGCAGGCCGUUAGUGCAAGCCCG ...((((..------((.((..((((((((....)))))))).)).))...))))(..(...(((.((((........)))).)))...)..)....... ( -28.70) >DroSim_CAF1 67882 94 + 1 UACGGAUUC------CGAAUUCCGGACAUAUACCUGUGUCCGUAUCCGUCUGUCCGUCCGUUCGGACUGCAUAAUUUAGCAGGCCGUUAGUGCAAGCCCG ...((((..------((.((..((((((((....)))))))).)).))...))))(..(...(((.((((........)))).)))...)..)....... ( -28.70) >DroEre_CAF1 71216 94 + 1 UCCGGAUUC------CGGAAUCCGGACAUAUACCUGUGUCCGCAUCCGUUUGUCCGUCCGCUCGGACUGCAUAAUUUAGCAGGCCUUUAGUGCAAGCCCA ..(((((..------((((...((((((((....))))))))..))))...)))))..((((.((.((((........)))).))...))))........ ( -32.40) >DroYak_CAF1 67911 100 + 1 UACGGAUUCGUUUUACGGAUUCCGGACAUAUACCUGUGUCCGUAUCCGUUUGUCCGUCCGUUCGGACUGCAUAAUUUAGCAGGCCGUUAGUGGAAGCCCG ..(((((.......((((((..((((((((....)))))))).))))))..)))))((((..(((.((((........)))).)))....))))...... ( -37.50) >consensus UACGGAUUC______CGAAUUCCGGACAUAUACCUGUGUCCGUAUCCGUCUGUCCGUCCGUUCGGACUGCAUAAUUUAGCAGGCCGUUAGUGCAAGCCCG .((((((...............((((((((....)))))))).))))))..(((((......)))))(((........)))(((...........))).. (-24.27 = -24.67 + 0.40)

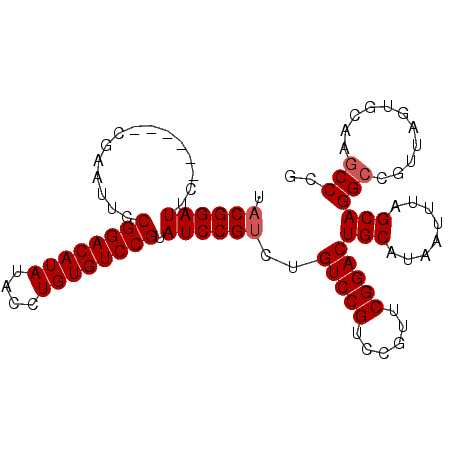
| Location | 13,467,156 – 13,467,250 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 92.95 |
| Mean single sequence MFE | -32.76 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.90 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13467156 94 - 22407834 CGGGUUUGCACUAACGGCCUGCUAAAUUAUGCAGUCCGAACGGACGGACAGACGGAUACGGACACAGGUAUAUGUCCGGAAUUCG------GAAUCCGUA (((((((((.(....))).(((........)))(((((......)))))...(((((.((((((........))))))..)))))------))))))).. ( -31.50) >DroSec_CAF1 66662 94 - 1 CGGGCUUGCACUAACGGCCUGCUAAAUUAUGCAGUCCGAACGGACGGACAGACGGAUACGGACACAGGUAUAUGUCCGGAAUUCG------GAAUCCGUA (((((..((.(....))).(((........)))))))).....(((((....(((((.((((((........))))))..)))))------...))))). ( -31.40) >DroSim_CAF1 67882 94 - 1 CGGGCUUGCACUAACGGCCUGCUAAAUUAUGCAGUCCGAACGGACGGACAGACGGAUACGGACACAGGUAUAUGUCCGGAAUUCG------GAAUCCGUA (((((..((.(....))).(((........)))))))).....(((((....(((((.((((((........))))))..)))))------...))))). ( -31.40) >DroEre_CAF1 71216 94 - 1 UGGGCUUGCACUAAAGGCCUGCUAAAUUAUGCAGUCCGAGCGGACGGACAAACGGAUGCGGACACAGGUAUAUGUCCGGAUUCCG------GAAUCCGGA .((((((.......))))))((........)).((((....))))(((....(((((.((((((........)))))).).))))------...)))... ( -33.80) >DroYak_CAF1 67911 100 - 1 CGGGCUUCCACUAACGGCCUGCUAAAUUAUGCAGUCCGAACGGACGGACAAACGGAUACGGACACAGGUAUAUGUCCGGAAUCCGUAAAACGAAUCCGUA .((....)).....(((.((((........)))).))).(((((((.....((((((.((((((........))))))..))))))....))..))))). ( -35.70) >consensus CGGGCUUGCACUAACGGCCUGCUAAAUUAUGCAGUCCGAACGGACGGACAGACGGAUACGGACACAGGUAUAUGUCCGGAAUUCG______GAAUCCGUA .(((((.........)))))((........)).(((((......)))))..((((((.((((((........))))))(....).........)))))). (-27.86 = -27.90 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:33 2006