| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,366,978 – 1,367,079 |
| Length | 101 |
| Max. P | 0.874458 |

| Location | 1,366,978 – 1,367,079 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
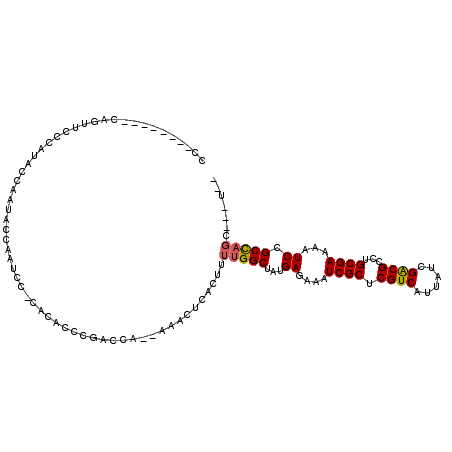
| Mean single sequence MFE | -17.52 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.73 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
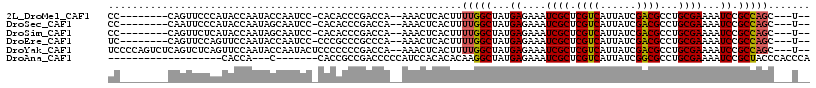
>2L_DroMel_CAF1 1366978 101 + 22407834 CC--------CAGUUCCCAUACCAAUACCAAUCC-CACACCCGACCA--AAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC---U-- ..--------...(((.((((((((.........-............--..........)))).)))).))).((((.((((......))))...))))............---.-- ( -15.98) >DroSec_CAF1 124000 101 + 1 CC--------CAAUUCCCAUACCAAUAGCAAUCC-CACACCCGACCA--AAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC---U-- ..--------...(((.((((((((.((......-............--.......)).)))).)))).))).((((.((((......))))...))))............---.-- ( -16.91) >DroSim_CAF1 128420 101 + 1 CC--------CAGUUCUCAUACCAAUAGCAAUCC-CACACCCGACCA--AAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC---U-- ..--------...((((((((((((.((......-............--.......)).)))).)))))))).((((.((((......))))...))))............---.-- ( -21.61) >DroEre_CAF1 142376 101 + 1 UC--------CAGUUCCAGUUCCAAUACCAAUCC-CCCGCCCGCCCA--AAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC---U-- ..--------........................-...((..(((((--(((.....))))))..........((((.((((......))))...))))......))..))---.-- ( -16.70) >DroYak_CAF1 127491 110 + 1 UCCCCAGUCUCAGUCUCAGUUCCAAUACCAAUACUCCCCCCCGACCA--AAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC---U-- .............(((((((......))..............(.(((--(((.....)))))))..)))))..((((.((((......))))...))))............---.-- ( -17.80) >DroAna_CAF1 150174 88 + 1 -------------------CACCA---C-------CACCGCCGACCCCCAUCCACACACAAGGCUAUGAGAAAUCGCUCGUCAUUAUCGGCGCCUGCGAAAAUCCGCUACCCACCCA -------------------.....---.-------...((((((.......((........))..(((((......))))).....))))))...(((......))).......... ( -16.10) >consensus CC________CAGUUCCCAUACCAAUACCAAUCC_CACACCCGACCA__AAACUCACUUUUGGCUAUGAGAAAUCGCUCGUCAUUAUCGACGCCUGCGAAAAUCCGCCAGC___U__ ...........................................................(((((...((....((((.((((......))))...))))...)).)))))....... (-14.68 = -14.73 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:53 2006