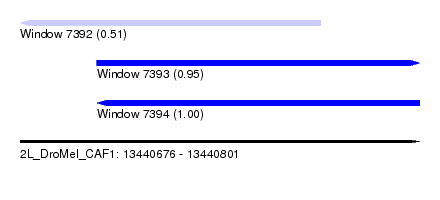
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,440,676 – 13,440,801 |
| Length | 125 |
| Max. P | 0.997079 |

| Location | 13,440,676 – 13,440,770 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -15.60 |
| Energy contribution | -17.18 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
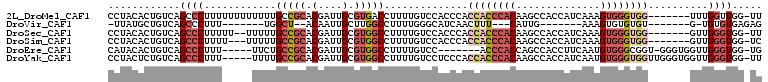
>2L_DroMel_CAF1 13440676 94 - 22407834 UUGUGGGUGGUGGGUGGACAAAAGGUCACGCAAUCGUGCGGCAAAAAAAAAAAAGGGCUGACAGUGUAGGGAGCGCAAGUUGGCCAAAUGUUUU ((((.((((((..((((((.....))).))).))))).).))))...........(((..((.((((.....))))..))..)))......... ( -23.10) >DroVir_CAF1 83664 81 - 1 AUG---AAAGUUGAUGCCCAAAAGGCCAAGCAAUUGU--AGCCA-------AAAGGGCUGACAGCAUAA-AAGCGAGACUUGGCAAAAUUACAU ...---..................(((((((...(((--((((.-------....)))).)))((....-..))..).)))))).......... ( -18.70) >DroSec_CAF1 47586 92 - 1 UUGUGGGUGGUGGGUGGACAAAAGGCCACGCAAUCGUGCGGCAAAAA--AAAAAGGGCUGACAGUGUAGGGAGCGCAAGUUGGCCAAAUGUUUU .((((.(...((.((((........)))).))..).)))).......--......(((..((.((((.....))))..))..)))......... ( -24.00) >DroSim_CAF1 48744 91 - 1 UUGUGGGUGGUGGGUGGACAAAAGGCCACGCAAUCGUGCGGCAAAAA---AAAAGGGCUGACAGUGUAGGGAGCGCAAGUUGGCCAAAUGUUUU .((((.(...((.((((........)))).))..).)))).......---.....(((..((.((((.....))))..))..)))......... ( -24.00) >DroEre_CAF1 52014 82 - 1 UGGUGGGU-------GGACAAAAGGCCACGCAAUCGUGCGGCAGAA-----AAAGGGCUGACAGUGUAUGGCGCGCAAGUUGGCCAAAUGUUUU ........-------(((((....(((.(((....))).)))....-----....(((..((.((((.....))))..))..)))...))))). ( -23.30) >DroYak_CAF1 48341 89 - 1 UUGUGGGUGGUGGGAGGACAAAAGGCCACGCAAUCGUGCGGCAAAA-----AAAGGGCUGACAGAGUAGGGAGCGCAAGUUGGCCAAAUGUUUU ..............((((((...(((((((((....))))......-----...(.(((..(......)..))).)....)))))...)))))) ( -21.70) >consensus UUGUGGGUGGUGGGUGGACAAAAGGCCACGCAAUCGUGCGGCAAAA_____AAAGGGCUGACAGUGUAGGGAGCGCAAGUUGGCCAAAUGUUUU ...............(((((....(((.(((....))).))).............(((..((.((((.....))))..))..)))...))))). (-15.60 = -17.18 + 1.58)


| Location | 13,440,700 – 13,440,801 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -16.54 |
| Energy contribution | -18.32 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13440700 101 + 22407834 CCUACACUGUCAGCCCUUUUUUUUUUUUGCCGCACGAUUGCGUGACCUUUUGUCCACCCACCACCCACAAGCCACCAUCAAAGUGGGUGG-------UUUGGUUGG-UU ........((((((..............))((((....)))))))).......(((.(((((((((((..............))))))))-------)..)).)))-.. ( -25.48) >DroVir_CAF1 83688 81 + 1 -UUAUGCUGUCAGCCCUUU-------UGGCU--ACAAUUGCUUGGCCUUUUGGGCAUCAACUUU---CAUUG-------AAAGUGUGUGU-------G-UGUGUGAGAG -((((((...((((((...-------.((((--(........)))))....))))....(((((---(...)-------)))))...)).-------.-.))))))... ( -22.30) >DroSec_CAF1 47610 99 + 1 CCUACACUGUCAGCCCUUUUU--UUUUUGCCGCACGAUUGCGUGGCCUUUUGUCCACCCACCACCCACAAGCCACCAUCAAAGUGGGUGG-------GUUGGGUGG-UU .....................--.....(((((........))))).......(((((((((((((((..............))))))))-------..)))))))-.. ( -34.14) >DroSim_CAF1 48768 98 + 1 CCUACACUGUCAGCCCUUUU---UUUUUGCCGCACGAUUGCGUGGCCUUUUGUCCACCCACCACCCACAAGCCACCAUCAAAGUGGGUGG-------GUUGGGUGG-UC ....................---.....(((((........))))).......(((((((((((((((..............))))))))-------..)))))))-.. ( -34.14) >DroEre_CAF1 52038 95 + 1 CAUACACUGUCAGCCCUUU-----UUCUGCCGCACGAUUGCGUGGCCUUUUGUCC-------ACCCACCAGCCACCUUCAAUGUGGGCGGU-GGGUGGUUGGGUGG-UG ....((((....((((...-----....(((((........))))).......((-------(((((((.(((..(......)..))))))-))))))..))))))-)) ( -39.50) >DroYak_CAF1 48365 103 + 1 CCUACUCUGUCAGCCCUUU-----UUUUGCCGCACGAUUGCGUGGCCUUUUGUCCUCCCACCACCCACAAGCCACCAUCAAUGUGGGUGGUUGGGUGGUUGGGUGG-UU .((((((............-----....(((((........))))).......((.(((((((((((((............))))))))).)))).))..))))))-.. ( -39.30) >consensus CCUACACUGUCAGCCCUUU_____UUUUGCCGCACGAUUGCGUGGCCUUUUGUCCACCCACCACCCACAAGCCACCAUCAAAGUGGGUGG_______GUUGGGUGG_UU ............((((............(((((.(....).)))))..............((((((((..............))))))))..........))))..... (-16.54 = -18.32 + 1.78)

| Location | 13,440,700 – 13,440,801 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -18.64 |
| Energy contribution | -20.56 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13440700 101 - 22407834 AA-CCAACCAAA-------CCACCCACUUUGAUGGUGGCUUGUGGGUGGUGGGUGGACAAAAGGUCACGCAAUCGUGCGGCAAAAAAAAAAAAGGGCUGACAGUGUAGG ..-(((.((..(-------((((((((..............))))))))).)))))(((....((((((((....))))((..............))))))..)))... ( -31.18) >DroVir_CAF1 83688 81 - 1 CUCUCACACA-C-------ACACACACUUU-------CAAUG---AAAGUUGAUGCCCAAAAGGCCAAGCAAUUGU--AGCCA-------AAAGGGCUGACAGCAUAA- ((.(((....-.-------..........(-------((((.---...))))).((((....((((((....))).--.))).-------...))))))).)).....- ( -17.90) >DroSec_CAF1 47610 99 - 1 AA-CCACCCAAC-------CCACCCACUUUGAUGGUGGCUUGUGGGUGGUGGGUGGACAAAAGGCCACGCAAUCGUGCGGCAAAAA--AAAAAGGGCUGACAGUGUAGG ..-(((((((((-------(((((((((.....)))))...))))))..)))))))(((...((((.((((....)))).......--......)))).....)))... ( -39.04) >DroSim_CAF1 48768 98 - 1 GA-CCACCCAAC-------CCACCCACUUUGAUGGUGGCUUGUGGGUGGUGGGUGGACAAAAGGCCACGCAAUCGUGCGGCAAAAA---AAAAGGGCUGACAGUGUAGG ..-(((((((((-------(((((((((.....)))))...))))))..)))))))(((...((((.((((....)))).......---.....)))).....)))... ( -39.12) >DroEre_CAF1 52038 95 - 1 CA-CCACCCAACCACCC-ACCGCCCACAUUGAAGGUGGCUGGUGGGU-------GGACAAAAGGCCACGCAAUCGUGCGGCAGAA-----AAAGGGCUGACAGUGUAUG ((-((((((..((((((-((((.((((.......)))).))))))))-------)).......(((.(((....))).)))....-----...))).))...))).... ( -41.50) >DroYak_CAF1 48365 103 - 1 AA-CCACCCAACCACCCAACCACCCACAUUGAUGGUGGCUUGUGGGUGGUGGGAGGACAAAAGGCCACGCAAUCGUGCGGCAAAA-----AAAGGGCUGACAGAGUAGG ..-.(((((..((.((((.(((((((((............))))))))))))).)).......(((.(((....))).)))....-----...))).)).......... ( -41.00) >consensus AA_CCACCCAAC_______CCACCCACUUUGAUGGUGGCUUGUGGGUGGUGGGUGGACAAAAGGCCACGCAAUCGUGCGGCAAAA_____AAAGGGCUGACAGUGUAGG ....(((((..........((((((((..............))))))))..............(((.(((....))).)))............))).)).......... (-18.64 = -20.56 + 1.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:22 2006