| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,435,515 – 13,435,619 |
| Length | 104 |
| Max. P | 0.960078 |

| Location | 13,435,515 – 13,435,619 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -20.46 |
| Energy contribution | -21.63 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
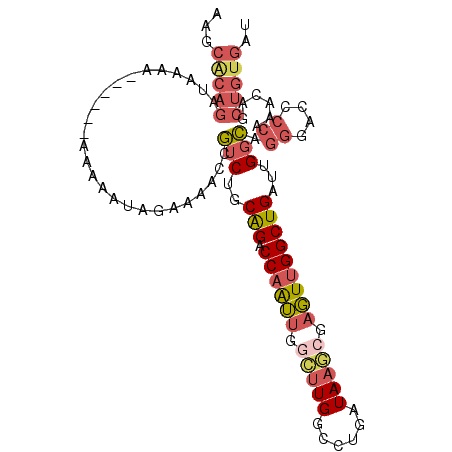
>2L_DroMel_CAF1 13435515 104 + 22407834 AAGCACAGAUAAAAAAAAUAAAAAAUAGAAAACCGUCUGCAGACCAAUUGGCUUGGCCUGAUAAGCGAGUUGGCUGAUUGGCAGGGACCCCAAACAGCUGUGAU ...(((((..........................(((..(((.((((((.(((((......))))).)))))))))...))).((....))......))))).. ( -30.50) >DroSec_CAF1 42428 99 + 1 AAGCACAGAUAAAAA-----AAAAAUAGAAAACCGUCUGCAGACCAGUUGGCUUGGCCUGAUAAGCGAGUUGGCUGAUUGGCAGGGAUCCCAAACAGCUGUGAU ...(((((.......-----..............(((..(((.(((..(.(((((......))))).)..))))))...))).((....))......))))).. ( -27.70) >DroSim_CAF1 43597 103 + 1 AAGCACAGAUAAAAAAUA-AAAAAAUAGAAAACCGUCUGCAGACCAAUUGGCUUGGCCUGAUAAGCGAGUUGGCUGAUUGGCAGGGACCCCAAACAGCUGUGAU ...(((((..........-...............(((..(((.((((((.(((((......))))).)))))))))...))).((....))......))))).. ( -30.50) >DroEre_CAF1 46962 94 + 1 AACCACAGA-A---------AAAAAUAGAAAGCCGUCUGCAGGCCAAUUGGCUUGGCCUGAUAAGCGAGUUGGCUGAUUGGCGGGGACCCCAAACAGCUGUGAU ...(((...-.---------..........(((((..((((((((((.....)))))))).))..)).)))(((((.((((.(....).)))).)))))))).. ( -35.70) >DroYak_CAF1 43302 95 + 1 AACCGCAGAUA---------AAAAAUAGAAAAUCGUCUGCAGUCCAAUUGGCUUGGCCUGAUAACACAGUUGGCUGAUUGGCAGGGACCCCAAACAGCUGUGAU ....((((((.---------..............))))))((.((((.....)))).)).....(((((((((((....))).((....))...)))))))).. ( -27.26) >DroPer_CAF1 54992 82 + 1 G-------GUAAAA------AAAAAGAGAA---CAACUUCGGACCAAGCCAAUUGGCCUGUUA-UCACCUGGGCUGACUGGU-----GCCCAAACAGCUGUGAU (-------((....------.....((.((---((.(....).....(((....))).)))).-))...(((((........-----)))))....)))..... ( -17.90) >consensus AAGCACAGAUAAAA______AAAAAUAGAAAACCGUCUGCAGACCAAUUGGCUUGGCCUGAUAAGCGAGUUGGCUGAUUGGCAGGGACCCCAAACAGCUGUGAU ...(((((..........................(((..(((.((((((.(((((......))))).)))))))))...))).((....))......))))).. (-20.46 = -21.63 + 1.17)


| Location | 13,435,515 – 13,435,619 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -18.92 |
| Energy contribution | -20.00 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
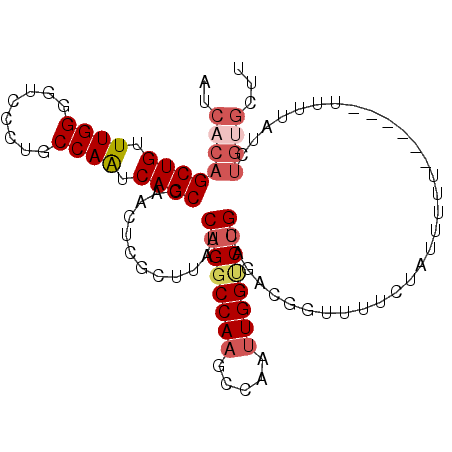
>2L_DroMel_CAF1 13435515 104 - 22407834 AUCACAGCUGUUUGGGGUCCCUGCCAAUCAGCCAACUCGCUUAUCAGGCCAAGCCAAUUGGUCUGCAGACGGUUUUCUAUUUUUUAUUUUUUUUAUCUGUGCUU ..((((((((.((((........)))).)))).(((.((((...((((((((.....)))))))).)).))))).......................))))... ( -25.60) >DroSec_CAF1 42428 99 - 1 AUCACAGCUGUUUGGGAUCCCUGCCAAUCAGCCAACUCGCUUAUCAGGCCAAGCCAACUGGUCUGCAGACGGUUUUCUAUUUUU-----UUUUUAUCUGUGCUU ..((((((((.((((........)))).)))).(((.((((...(((((((.......))))))).)).)))))..........-----........))))... ( -23.80) >DroSim_CAF1 43597 103 - 1 AUCACAGCUGUUUGGGGUCCCUGCCAAUCAGCCAACUCGCUUAUCAGGCCAAGCCAAUUGGUCUGCAGACGGUUUUCUAUUUUUU-UAUUUUUUAUCUGUGCUU ..((((((((.((((........)))).)))).(((.((((...((((((((.....)))))))).)).)))))...........-...........))))... ( -25.60) >DroEre_CAF1 46962 94 - 1 AUCACAGCUGUUUGGGGUCCCCGCCAAUCAGCCAACUCGCUUAUCAGGCCAAGCCAAUUGGCCUGCAGACGGCUUUCUAUUUUU---------U-UCUGUGGUU ((((((((((.((((((...)).)))).))))......(((..(((((((((.....)))))))).)...)))...........---------.-..)))))). ( -31.40) >DroYak_CAF1 43302 95 - 1 AUCACAGCUGUUUGGGGUCCCUGCCAAUCAGCCAACUGUGUUAUCAGGCCAAGCCAAUUGGACUGCAGACGAUUUUCUAUUUUU---------UAUCUGCGGUU ..((((((((.((((........)))).))))....))))......(((...))).....(((((((((..(...........)---------..))))))))) ( -26.30) >DroPer_CAF1 54992 82 - 1 AUCACAGCUGUUUGGGC-----ACCAGUCAGCCCAGGUGA-UAACAGGCCAAUUGGCUUGGUCCGAAGUUG---UUCUCUUUUU------UUUUAC-------C ...((((((..((((((-----....((((.(....))))-)..((((((....)))))))))))))))))---).........------......-------. ( -24.30) >consensus AUCACAGCUGUUUGGGGUCCCUGCCAAUCAGCCAACUCGCUUAUCAGGCCAAGCCAAUUGGUCUGCAGACGGUUUUCUAUUUUU______UUUUAUCUGUGCUU ..((((((((.((((........)))).))))............((((((((.....))))))))................................))))... (-18.92 = -20.00 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:18 2006