| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,434,730 – 13,434,837 |
| Length | 107 |
| Max. P | 0.838629 |

| Location | 13,434,730 – 13,434,837 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.62 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13434730 107 + 22407834 UUAUUUAUUUUCCUGUUAAAUCCCCUCGACCAAUAUCCUGAAUCAAGGAUAUUAACGGGUUACGAGUCUAAGAGUACACAUUCCACUAACUGAUUAGUGUGGGAAAU---------- ....................(((((((((((((((((((......))))))))....)))..)))).................((((((....)))))).))))...---------- ( -23.90) >DroSec_CAF1 41637 117 + 1 UUAUUUAUUUUCCUGUUAAAUCCCCUCGACCAAUAUCCUGAAUCAAGGAUAUUAACGGGUUACGAGCCUAAGAGUACACAUUACACUAACUGAUUAGUGCGGGAAAUACCUAAGUAU .((((((.(((((((.........(((....((((((((......))))))))...((((.....))))..))).........((((((....)))))))))))))....)))))). ( -27.80) >DroSim_CAF1 42776 117 + 1 UUAUUUAUUUUCCUGUUAAAACCCCUCGACCAAUAUCCUGAAUCAAGGAUAUUAACGGGUUACGAGUCUAAGAGUACACAUUCCACUAACUGAUUAGUGCCGGAAAUACCUAAGUCU ........(((((...........(((((((((((((((......))))))))....)))..)))).................((((((....))))))..)))))........... ( -22.60) >DroEre_CAF1 46205 113 + 1 UUAUUUAUUUUCCUGUUAAGGCCCCACGACUAAUAUCCCGAAUAAAGGAUAUUAGCGGGUUACGCUUCUAAGAGUAUCAAUUCUAGAAACUGAUUAGUUUGGGAUAU----AGUUCU ...........(((....)))......(((((.((((((((((...........(((.....)))(((((.((((....)))))))))........)))))))))))----)))).. ( -26.80) >DroYak_CAF1 42483 111 + 1 UUAUUUAUUUUCCCGUUGAAGCCCCUCGGCCCAUAUCCUGAAUCAAGGAUAUUAAAGGGUUAAGAGAUAUAUGGUAGCAAG--UAGUAACUGAUUUGUAUGGGAGAU----AAGUUU ..((((((((.(((((........((..(((((((((((......)))))))....))))..))............(((((--(........)))))))))))))))----)))).. ( -29.50) >consensus UUAUUUAUUUUCCUGUUAAAGCCCCUCGACCAAUAUCCUGAAUCAAGGAUAUUAACGGGUUACGAGUCUAAGAGUACACAUUCCACUAACUGAUUAGUGUGGGAAAU____AAGUCU ........((((((((........(((((((((((((((......))))))))....)))..)))).((((.(((.............)))..)))).))))))))........... (-16.38 = -16.62 + 0.24)


| Location | 13,434,730 – 13,434,837 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
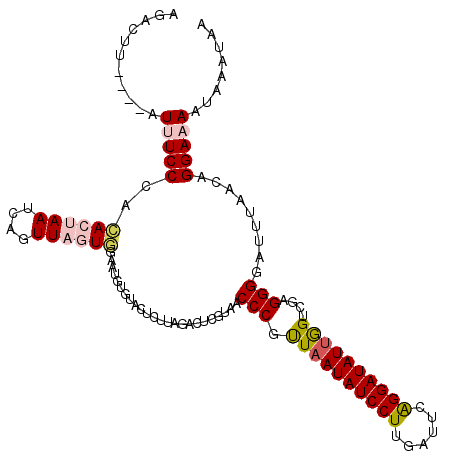
>2L_DroMel_CAF1 13434730 107 - 22407834 ----------AUUUCCCACACUAAUCAGUUAGUGGAAUGUGUACUCUUAGACUCGUAACCCGUUAAUAUCCUUGAUUCAGGAUAUUGGUCGAGGGGAUUUAACAGGAAAAUAAAUAA ----------.(((((..((((((....))))))...(((....(((....((((..(((....((((((((......))))))))))))))).)))....))))))))........ ( -25.70) >DroSec_CAF1 41637 117 - 1 AUACUUAGGUAUUUCCCGCACUAAUCAGUUAGUGUAAUGUGUACUCUUAGGCUCGUAACCCGUUAAUAUCCUUGAUUCAGGAUAUUGGUCGAGGGGAUUUAACAGGAAAAUAAAUAA ...((.((((((.....(((((((....))))))).....)))).)).))........((.(((((..((((((((.(((....)))))))))))...))))).))........... ( -28.50) >DroSim_CAF1 42776 117 - 1 AGACUUAGGUAUUUCCGGCACUAAUCAGUUAGUGGAAUGUGUACUCUUAGACUCGUAACCCGUUAAUAUCCUUGAUUCAGGAUAUUGGUCGAGGGGUUUUAACAGGAAAAUAAAUAA ...........(((((..((((((....))))))...(((..((((((.(((.((.....)).(((((((((......)))))))))))).))))))....))))))))........ ( -28.30) >DroEre_CAF1 46205 113 - 1 AGAACU----AUAUCCCAAACUAAUCAGUUUCUAGAAUUGAUACUCUUAGAAGCGUAACCCGCUAAUAUCCUUUAUUCGGGAUAUUAGUCGUGGGGCCUUAACAGGAAAAUAAAUAA ......----...(((((.........(((((((((........)).)))))))......((((((((((((......)))))))))).)))))))(((....)))........... ( -28.10) >DroYak_CAF1 42483 111 - 1 AAACUU----AUCUCCCAUACAAAUCAGUUACUA--CUUGCUACCAUAUAUCUCUUAACCCUUUAAUAUCCUUGAUUCAGGAUAUGGGCCGAGGGGCUUCAACGGGAAAAUAAAUAA ......----...((((.........(((.....--...)))........((((((..(((....(((((((......))))))))))..)))))).......)))).......... ( -22.50) >consensus AGACUU____AUUUCCCACACUAAUCAGUUAGUGGAAUGUGUACUCUUAGACUCGUAACCCGUUAAUAUCCUUGAUUCAGGAUAUUGGUCGAGGGGAUUUAACAGGAAAAUAAAUAA ...........(((((..((((((....))))))........................(((.((((((((((......))))))))))....))).........)))))........ (-18.10 = -18.98 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:16 2006