
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
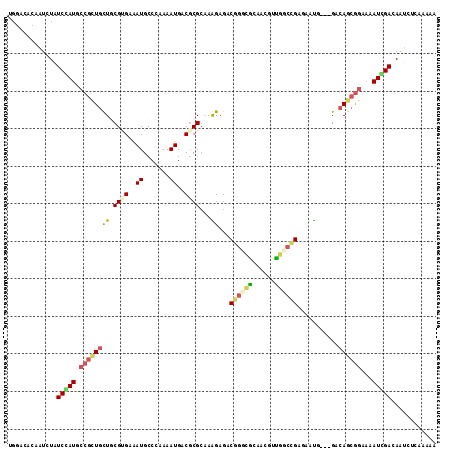
| Location | 13,405,579 – 13,405,693 |
| Length | 114 |
| Max. P | 0.777183 |

| Location | 13,405,579 – 13,405,693 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.52 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -16.53 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13405579 114 + 22407834 CGGCCCCAAUCUAUCGAUGCCGCUGCUGCGCGAGAUGCCCAAAAUGACGCGACAGGAGACAGGUGGACCCUUGGCUGAAAACAGCGGACAGCGGAAGAUUGAAAAUCUAAAAAA ......((((((.(((.(((((((((((((((..((.......))..)))).))).((.((((......)))).)).....)))))).)).))).))))))............. ( -33.70) >DroVir_CAF1 19274 111 + 1 CACACAGAAUCUAUCCAUGCCGCUGCUACGAGAAAUGCCAAAAAUGACGCGCAAAGAGACGGCCACAAAGUUGGACGGAAAUC---GUCAGCAGAAAAUGGAUAAUCUCAAGAA ......((...(((((((....(((((((((....((((((...((..((.(........)))..))...)))).))....))---)).)))))...)))))))...))..... ( -27.40) >DroPse_CAF1 47132 111 + 1 UGGGCAUAAUCUAUCGAUGCCGCUGCUGCGUGAAAUGCCCAAAAUGACGCGCAAAGAGACGGGCGCCACAUUGCCCGUGAAUG---GACAGCGGGGCAUCGACAGUCUCAAAAA ((((......((.((((((((.((((((((((..((.......))..)))))......(((((((......))))))).....---...))))))))))))).)).)))).... ( -42.20) >DroGri_CAF1 15819 111 + 1 UACACCGAAUUUAUCCAUGCCGCUGCUGCGCGAAAUGCCUAAGAUGACGCGCAAGGAGACGGCUGCAACGCCCGCCGAGACUC---GACAUCGUAAAAUGGAUAAUCUCAAGAA ......((..((((((((((((((.(((((((..((.......))..))))).)).)).))))....(((...(.((.....)---).)..)))...))))))))..))..... ( -32.10) >DroAna_CAF1 68752 111 + 1 GGGUCACAAUCUGUCCAUGCCAUUACUUCGUGAAAUGCCCAAAAUGACCCGCCAGGAGACGGGUACAACGCUGGCGGGGAAUG---GCCAGCGGAAAAUCGACAACCUUAAAAA ((((((.....((..(((..(((......)))..)))..))...))))))(((.(....).)))....(((((((.(....).---)))))))..................... ( -33.50) >DroPer_CAF1 21711 111 + 1 UGGGCAUAAUCUAUCGAUGCCGCUGUUGCGUGAAAUGCCCAAAAUGACGCGCAAAGAGACGGGCGCCACAUUGCCCGUGAAUG---GACAGCGGGGCAUCGACAGUCUCAAAAA ((((......((.(((((((((((((((((((..((.......))..)))))......(((((((......))))))).....---))))))..)))))))).)).)))).... ( -42.10) >consensus UGGACACAAUCUAUCCAUGCCGCUGCUGCGUGAAAUGCCCAAAAUGACGCGCAAAGAGACGGGCGCAACGUUGGCCGAGAAUG___GACAGCGGAAAAUCGACAAUCUCAAAAA .............(((((.((((((((.((((..((.......))..))))...))...((((((......))))))...........))))))...)))))............ (-16.53 = -16.73 + 0.20)


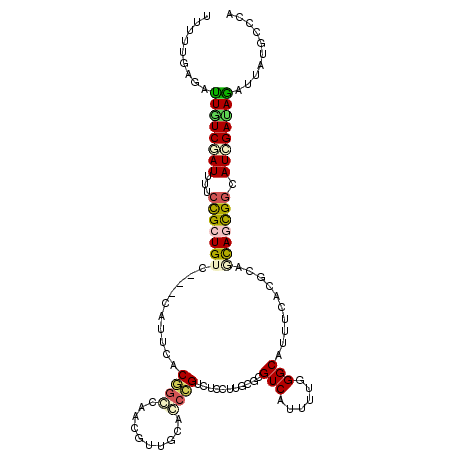
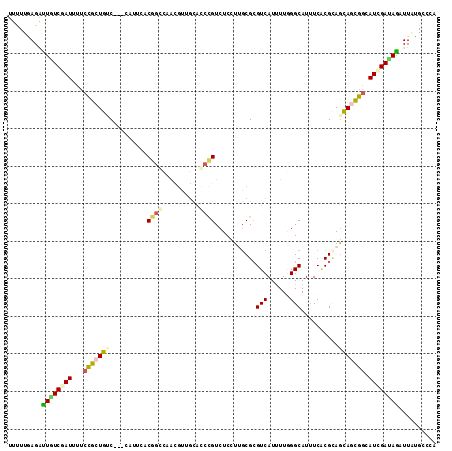
| Location | 13,405,579 – 13,405,693 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.52 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -18.22 |
| Energy contribution | -18.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13405579 114 - 22407834 UUUUUUAGAUUUUCAAUCUUCCGCUGUCCGCUGUUUUCAGCCAAGGGUCCACCUGUCUCCUGUCGCGUCAUUUUGGGCAUCUCGCGCAGCAGCGGCAUCGAUAGAUUGGGGCCG ..........(..((((((..((.((.(((((((.....((..((((..(....)..))))...)).......((.((.....)).))))))))))).))..))))))..)... ( -34.90) >DroVir_CAF1 19274 111 - 1 UUCUUGAGAUUAUCCAUUUUCUGCUGAC---GAUUUCCGUCCAACUUUGUGGCCGUCUCUUUGCGCGUCAUUUUUGGCAUUUCUCGUAGCAGCGGCAUGGAUAGAUUCUGUGUG .....(((.(((((((((..((((((.(---((.....(.((((....((((((((......))).)))))..))))).....)))))))))..).)))))))).)))...... ( -31.50) >DroPse_CAF1 47132 111 - 1 UUUUUGAGACUGUCGAUGCCCCGCUGUC---CAUUCACGGGCAAUGUGGCGCCCGUCUCUUUGCGCGUCAUUUUGGGCAUUUCACGCAGCAGCGGCAUCGAUAGAUUAUGCCCA .........((((((((((((.(((((.---.....((((((........)))))).....(((.((......)).)))......))))).).))))))))))).......... ( -42.70) >DroGri_CAF1 15819 111 - 1 UUCUUGAGAUUAUCCAUUUUACGAUGUC---GAGUCUCGGCGGGCGUUGCAGCCGUCUCCUUGCGCGUCAUCUUAGGCAUUUCGCGCAGCAGCGGCAUGGAUAAAUUCGGUGUA ...(((((.((((((((....(((((((---..((....)).)))))))..((((.((.((.(((((..(((....).))..))))))).)))))))))))))).))))).... ( -37.50) >DroAna_CAF1 68752 111 - 1 UUUUUAAGGUUGUCGAUUUUCCGCUGGC---CAUUCCCCGCCAGCGUUGUACCCGUCUCCUGGCGGGUCAUUUUGGGCAUUUCACGAAGUAAUGGCAUGGACAGAUUGUGACCC .......(((((.(((((((((((((((---........))))))((..((((((((....))))))).((((((.........)))))).)..))..))).))))))))))). ( -39.80) >DroPer_CAF1 21711 111 - 1 UUUUUGAGACUGUCGAUGCCCCGCUGUC---CAUUCACGGGCAAUGUGGCGCCCGUCUCUUUGCGCGUCAUUUUGGGCAUUUCACGCAACAGCGGCAUCGAUAGAUUAUGCCCA .........(((((((((((..((((((---((...(((.((((.(.(((....))).).)))).))).....)))((.......)).)))))))))))))))).......... ( -41.30) >consensus UUUUUGAGAUUGUCGAUUUUCCGCUGUC___CAUUCACGGCCAACGUUGCACCCGUCUCCUUGCGCGUCAUUUUGGGCAUUUCACGCAGCAGCGGCAUCGAUAGAUUAUGCCCA .........((((((((...(((((((..........((((..........))))...........(((......)))..........))))))).)))))))).......... (-18.22 = -18.42 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:12 2006