| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,399,541 – 13,399,648 |
| Length | 107 |
| Max. P | 0.965935 |

| Location | 13,399,541 – 13,399,648 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.83 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -27.17 |
| Energy contribution | -27.82 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13399541 107 + 22407834 AGCUGCUCGUCGGCAGCGCUCGUCCGUAUAAGUCGCCAGAUGCGUCGAUG---CGACGCAAUUUGGAGAACGAGCGGCCGCUCAGUGAGAACCGCUUCCCAAUUCAUCGU------- .((((((....))))))((((((.........((.((((((((((((...---)))))).)))))).))))))))((...(((...)))..)).................------- ( -37.40) >DroVir_CAF1 11896 111 + 1 AAUUGCUUAUUGGUAGCGCGCGACAAUACAAAUCGGCUAAUGCGUCG------CAGCGUAAUUUAGAAAGCGAGCGGCCGCAAAGCGAGAAUCGUUUGCCAAUUGGUCGUCGUACAG ..((((((.((((..(((((((((....(.....)((....))))))------).))))...)))).))))))(((((((((((.((.....))))))).....))))))....... ( -33.70) >DroPse_CAF1 15767 107 + 1 AGCUGCUCGUUGGAAGCGCCCGUCAAUACAAAUCGCCAGAUGUGUCGUCG---CGACGCAAUUUGGAAAGCGGGCGACCGCUAAGUGAGAAUCGCUUGCCAAUCCAACGA------- ......((((((((..(((((((............((((((((((((...---)))))).))))))...)))))))...((.(((((.....)))))))...))))))))------- ( -42.46) >DroWil_CAF1 12298 116 + 1 AACUGCUCGUUGGCAGCGCUCGACCGUACAAAUCUCCAAAUGCGUCUCUGGAGGAACGCAAUUUGGAAAGCGGGCGACCGCUAAGCGAGAAUCGUUUGCCAAUUAGGCGGCGAACC- ..(((((.((((((((((((((............(((((((((((..(....)..)))).))))))).(((((....)))))...))))...)).))))))))..)))))......- ( -45.80) >DroAna_CAF1 63073 107 + 1 AGCUGCUCAUAGGCAGCGCCCGUCAGUACAAGUCGCCAAAUGAGACUCUA---CGACGCAAUUUGGAUAGCGGGCGACCGCUAAGUGAGAAUCGCUUGCCAAUCCGGCGA------- .((((((....))))))(((((((.(((..((((.........)))).))---)))))......(((((((((....)))))(((((.....)))))....)))))))..------- ( -41.50) >DroPer_CAF1 15444 107 + 1 AGCUGCUCGUUGGAAGCGCCCGUCAAUACAAAUCGCCAGAUGUGUCGUCG---CGACGCAAUUUGGAAAGCGGGCGACCGCUAAGUGAGAAUCGCUUGCCAAUCCGACGA------- ......((((((((..(((((((............((((((((((((...---)))))).))))))...)))))))...((.(((((.....)))))))...))))))))------- ( -42.16) >consensus AGCUGCUCGUUGGCAGCGCCCGUCAAUACAAAUCGCCAAAUGCGUCGCUG___CGACGCAAUUUGGAAAGCGGGCGACCGCUAAGUGAGAAUCGCUUGCCAAUCCGACGA_______ .((((((....))))))...((((...........((((((((((((......)))))).))))))...((((((((((((...))).)..))))))))......))))........ (-27.17 = -27.82 + 0.64)


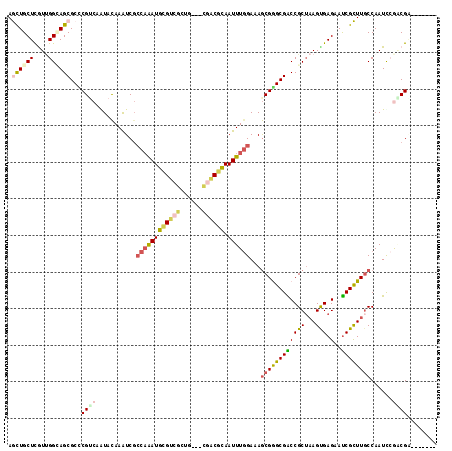
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:10 2006