
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,394,871 – 13,394,968 |
| Length | 97 |
| Max. P | 0.593587 |

| Location | 13,394,871 – 13,394,968 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.70 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -16.17 |
| Energy contribution | -15.53 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13394871 97 - 22407834 ---C--UGCUAAU---GA-GA--GUGUU-UGUAUUUACAGAAUUGGCUUGUGCGCAUUCAGCG-GGUACAAAAUCUACCCCGGUCAUGGCAAGACCAUGGUCAAGAUCGA ---.--.((((((---(.-((--((.((-(((....)))))....)))).....)))).)))(-((((.......))))).(..(((((.....)))))..)........ ( -25.40) >DroVir_CAF1 9603 100 - 1 CCUA--UGCUC-----AAACAAUACGU--UUUUAUUGCAGAAUCGGUUUGUGUGCAUUCAGCG-GGUAUAAGAUUUACCCCGGCCAUGGCAAGACCAUGGUCAAAAUCGA ...(--(((..-----..((((..((.--((((.....)))).))..))))..))))...(.(-((((.......))))))((((((((.....))))))))........ ( -27.90) >DroPse_CAF1 8143 104 - 1 CC-A--GCCUAAUAAUGAAUA--AUGUAAUCUCCUUACAGAAUUGGCUUGUGUGCAUUCAGCG-GGUACAAAAUCUACCCCGGCCAUGGCAAGACCAUGGUCAAGAUUGA .(-(--..((.....(((((.--.(((((.....)))))......((......)))))))(.(-((((.......))))))((((((((.....)))))))).))..)). ( -26.40) >DroWil_CAF1 10236 101 - 1 GA-AAUUUAUACU---AA-AA--UUCUA-UUGUUUUGCAGAAUCGGUUUGUGUUCAUUCAGUG-GGUACAAAAUCUACCCCGGCCAUGGCAAGACCAUGGUUAAGGUUGA ..-.........(---((-((--(....-..)))))).........((((((..(((...)))-..)))))).((.(((..((((((((.....))))))))..))).)) ( -23.70) >DroMoj_CAF1 9054 100 - 1 ACUA--UGCUU-----AAACAAAACAU--UUUAUUUGCAGAAUUGGCUUGUGUGCAUUCAGUG-GGUACAAAAUCUACCCCGGCCAUGGCAAGACCAUGGUCAAAGUCGA ((((--(((..-----..((((..((.--(((.......))).))..))))..))))..)))(-((((.......))))).((((((((.....))))))))........ ( -26.90) >DroPer_CAF1 8194 105 - 1 CC-C--AGCUAAUAAUGAAUA--AUGUAAUCUCCUUACAGAAUUGGCUUGUGUGCAUUCAGCGGGGUACAAAAUCUACCCCGGCCAUGGCAAGACCAUGGUCAAGAUUGA ..-.--.(((...........--.(((((.....)))))((((..((......)))))))))((((((.......))))))((((((((.....))))))))........ ( -31.20) >consensus CC_A__UGCUAAU___AAACA__AUGUA_UUUAUUUACAGAAUUGGCUUGUGUGCAUUCAGCG_GGUACAAAAUCUACCCCGGCCAUGGCAAGACCAUGGUCAAGAUCGA .......................................((((..((....))..))))...(.((((.......)))).)((((((((.....))))))))........ (-16.17 = -15.53 + -0.64)

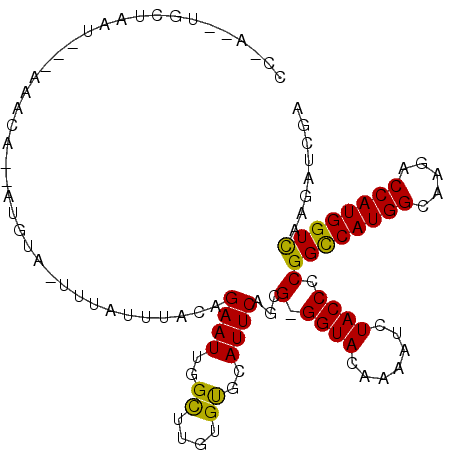
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:07 2006